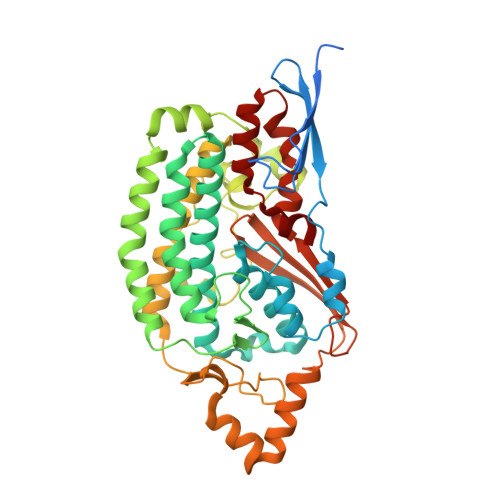
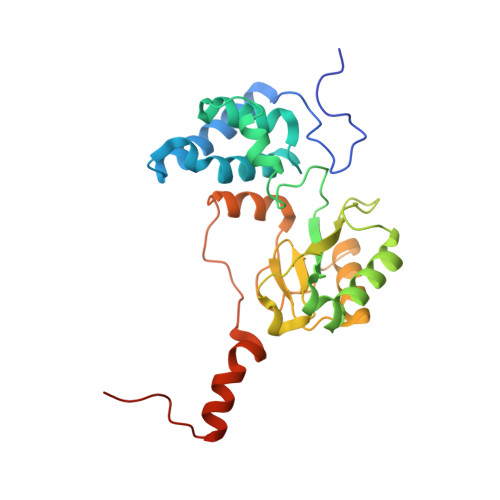
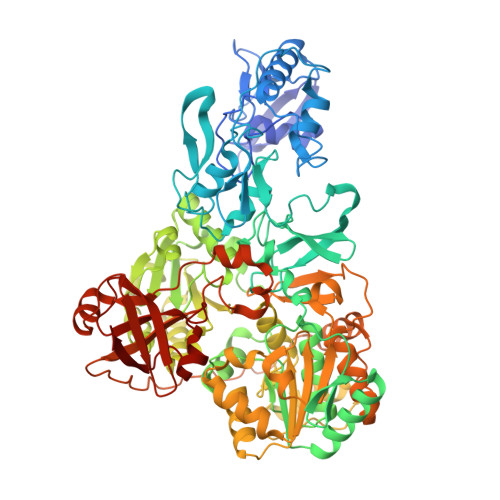
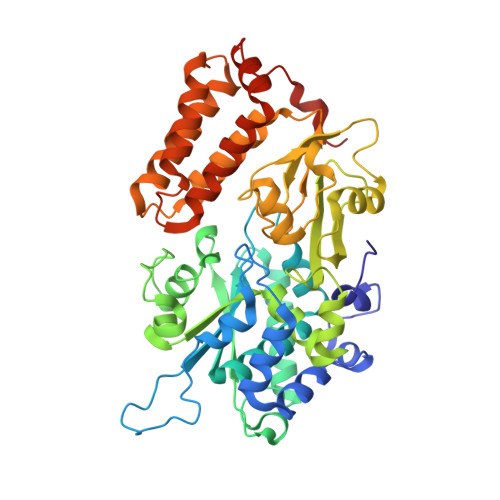
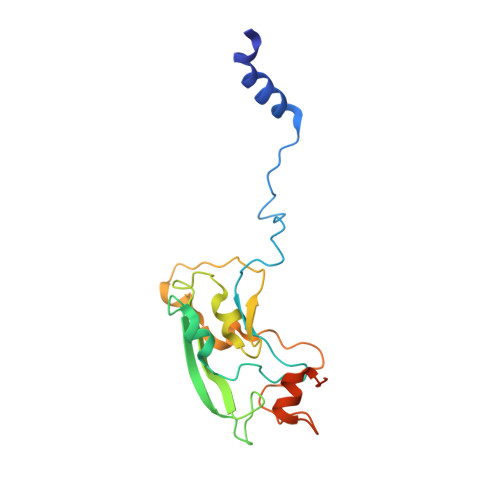
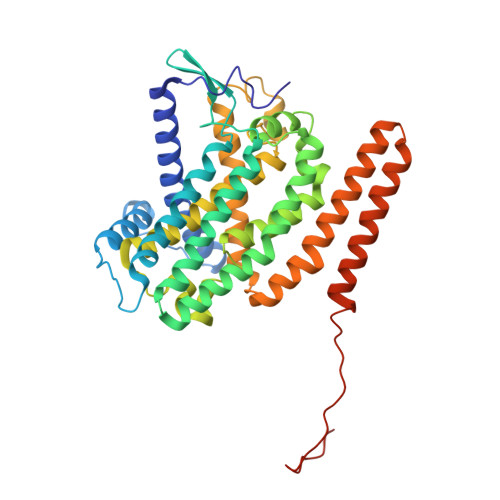
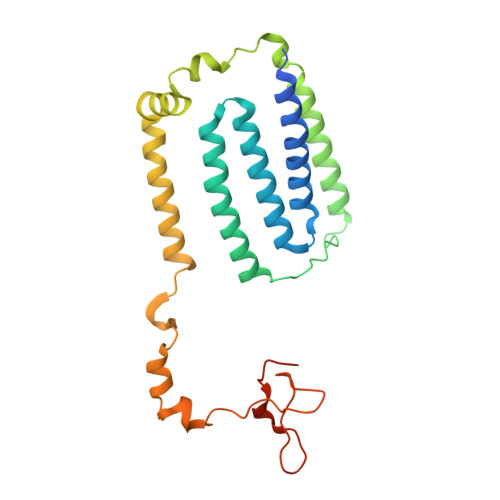
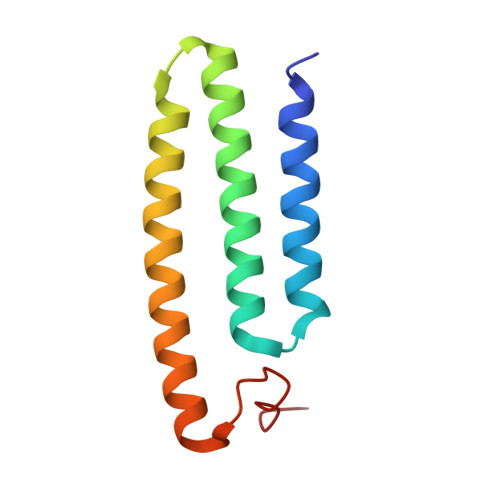
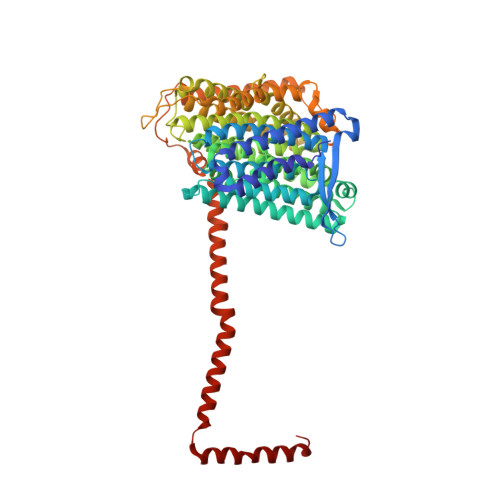
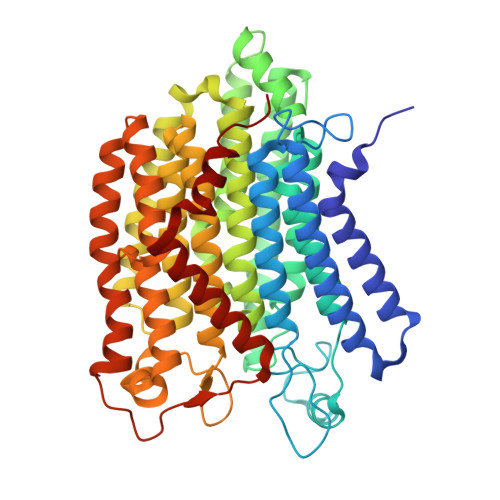
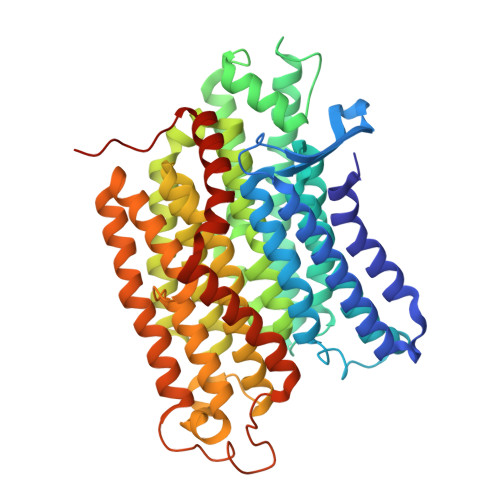
Structure of mycobacterial respiratory complex I.
Liang, Y., Plourde, A., Bueler, S.A., Liu, J., Brzezinski, P., Vahidi, S., Rubinstein, J.L.(2023) Proc Natl Acad Sci U S A 120: e2214949120-e2214949120
- PubMed: 36952383
- DOI: https://doi.org/10.1073/pnas.2214949120
- Primary Citation of Related Structures:
8E9G, 8E9H, 8E9I - PubMed Abstract:
Oxidative phosphorylation, the combined activity of the electron transport chain (ETC) and adenosine triphosphate synthase, has emerged as a valuable target for the treatment of infection by Mycobacterium tuberculosis and other mycobacteria. The mycobacterial ETC is highly branched with multiple dehydrogenases transferring electrons to a membrane-bound pool of menaquinone and multiple oxidases transferring electrons from the pool. The proton-pumping type I nicotinamide adenine dinucleotide (NADH) dehydrogenase (Complex I) is found in low abundance in the plasma membranes of mycobacteria in typical in vitro culture conditions and is often considered dispensable. We found that growth of Mycobacterium smegmatis in carbon-limited conditions greatly increased the abundance of Complex I and allowed isolation of a rotenone-sensitive preparation of the enzyme. Determination of the structure of the complex by cryoEM revealed the "orphan" two-component response regulator protein MSMEG_2064 as a subunit of the assembly. MSMEG_2064 in the complex occupies a site similar to the proposed redox-sensing subunit NDUFA9 in eukaryotic Complex I. An apparent purine nucleoside triphosphate within the NuoG subunit resembles the GTP-derived molybdenum cofactor in homologous formate dehydrogenase enzymes. The membrane region of the complex binds acyl phosphatidylinositol dimannoside, a characteristic three-tailed lipid from the mycobacterial membrane. The structure also shows menaquinone, which is preferentially used over ubiquinone by gram-positive bacteria, in two different positions along the quinone channel, comparable to ubiquinone in other structures and suggesting a conserved quinone binding mechanism.
- Molecular Medicine Program, The Hospital for Sick Children, Toronto M5G 0A4, Canada.
Organizational Affiliation: