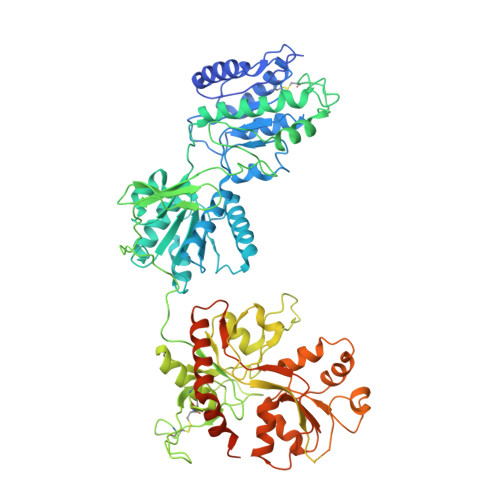
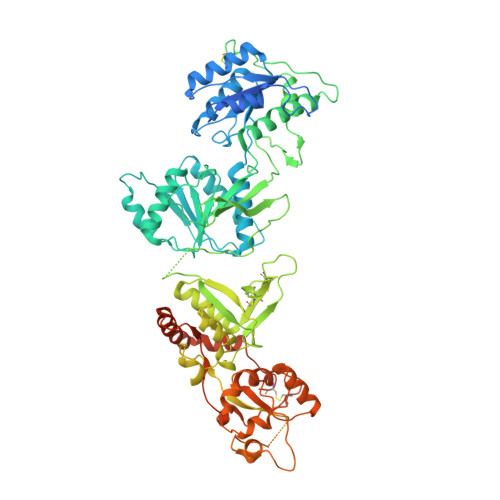
Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Chou, T.H., Kang, H., Simorowski, N., Traynelis, S.F., Furukawa, H.(2022) Mol Cell 82: 4548
- PubMed: 36309015
- DOI: https://doi.org/10.1016/j.molcel.2022.10.008
- Primary Citation of Related Structures:
8E92, 8E93, 8E94, 8E96, 8E97, 8E98, 8E99 - PubMed Abstract:
Neurotransmission mediated by diverse subtypes of N-methyl-D-aspartate receptors (NMDARs) is fundamental for basic brain functions and development as well as neuropsychiatric diseases and disorders. NMDARs are glycine- and glutamate-gated ion channels that exist as heterotetramers composed of obligatory GluN1 and GluN2(A-D) and/or GluN3(A-B). The GluN2C and GluN2D subunits form ion channels with distinct properties and spatio-temporal expression patterns. Here, we provide the structures of the agonist-bound human GluN1-2C NMDAR in the presence and absence of the GluN2C-selective positive allosteric potentiator (PAM), PYD-106, the agonist-bound GluN1-2A-2C tri-heteromeric NMDAR, and agonist-bound GluN1-2D NMDARs by single-particle electron cryomicroscopy. Our analysis shows unique inter-subunit and domain arrangements of the GluN2C NMDARs, which contribute to functional regulation and formation of the PAM binding pocket and is distinct from GluN2D NMDARs. Our findings here provide the fundamental blueprint to study GluN2C- and GluN2D-containing NMDARs, which are uniquely involved in neuropsychiatric disorders.
- W.M. Keck Structural Biology Laboratory, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724, USA.
Organizational Affiliation: