Leveraging the Structure of DNAJA1 to Discover Novel Potential Pancreatic Cancer Therapies.
Roth, H.E., De Lima Leite, A., Palermo, N.Y., Powers, R.(2022) Biomolecules 12
- PubMed: 36291603
- DOI: https://doi.org/10.3390/biom12101391
- Primary Citation of Related Structures:
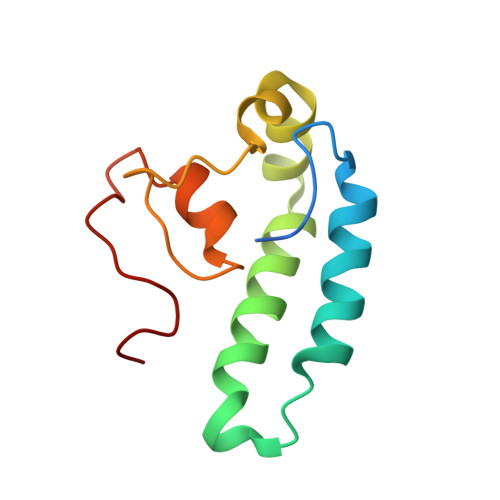
8E2O - PubMed Abstract:
Pancreatic cancer remains one of the deadliest forms of cancer with a 5-year survival rate of only 11%. Difficult diagnosis and limited treatment options are the major causes of the poor outcome for pancreatic cancer. The human protein DNAJA1 has been proposed as a potential therapeutic target for pancreatic cancer, but its cellular and biological functions remain unclear. Previous studies have suggested that DNAJA1's cellular activity may be dependent upon its protein binding partners. To further investigate this assertion, the first 107 amino acid structures of DNAJA1 were solved by NMR, which includes the classical J-domain and its associated linker region that is proposed to be vital to DNAJA1 functionality. The DNAJA1 NMR structure was then used to identify both protein and ligand binding sites and potential binding partners that may suggest the intracellular roles of DNAJA1. Virtual drug screenings followed by NMR and isothermal titration calorimetry identified 5 drug-like compounds that bind to two different sites on DNAJA1. A pull-down assay identified 8 potentially novel protein binding partners of DNAJA1. These proteins in conjunction with our previously published metabolomics study support a vital role for DNAJA1 in cellular oncogenesis and pancreatic cancer.
- Department of Chemistry, University of Nebraska-Lincoln, Lincoln, NE 68588, USA.
Organizational Affiliation: