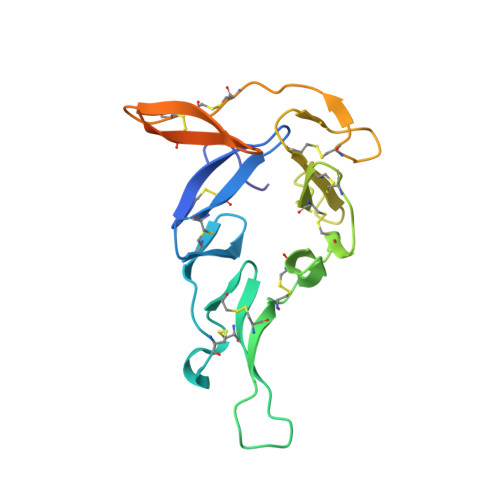
Structural analysis of Plasmodium falciparum ookinete surface antigen Pfs28 relevant for malaria vaccine design.
Shukla, N., Tang, W.K., Tolia, N.H.(2022) Sci Rep 12: 19556-19556
- PubMed: 36379968
- DOI: https://doi.org/10.1038/s41598-022-24054-3
- Primary Citation of Related Structures:
8E1Z - PubMed Abstract:
Pfs28 is a Plasmodium falciparum malaria transmission-blocking vaccine candidate that is anchored to the parasite surface through a C-terminal glycosylphosphatidylinositol (GPI) moiety, and plays a role in parasite survival in the mosquito midgut. Pfs28 contains epidermal growth factor (EGF)-like domains and is part of a family of sexual stage malaria proteins that includes the related vaccine antigen Pfs25. The lack of structural definition of Pfs28 and the immune response to this candidate has limited further malaria vaccine development for this antigen. Here, we present the crystal structure of Pfs28, examine its conservation with P. vivax Pvs28, and evaluate the cross-reactivity of Pfs28 to antibodies that recognize Pfs25. Pfs28 is comprised of four EGF-like domains stabilized by ten disulfide bridges with an overall architecture that highly resembles Pfs25. Despite the high sequence and structural similarity between these antigens, no cross reactivity of Pfs28 to anti-Pfs25 monoclonal antibodies could be demonstrated.
- Host-Pathogen Interactions and Structural Vaccinology Section, Laboratory of Malaria Immunology and Vaccinology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, USA.
Organizational Affiliation: