Structure-Based Study to Overcome Cross-Reactivity of Novel Androgen Receptor Inhibitors.
Radaeva, M., Li, H., LeBlanc, E., Dalal, K., Ban, F., Ciesielski, F., Chow, B., Morin, H., Awrey, S., Singh, K., Rennie, P.S., Lallous, N., Cherkasov, A.(2022) Cells 11
- PubMed: 36139361
- DOI: https://doi.org/10.3390/cells11182785
- Primary Citation of Related Structures:
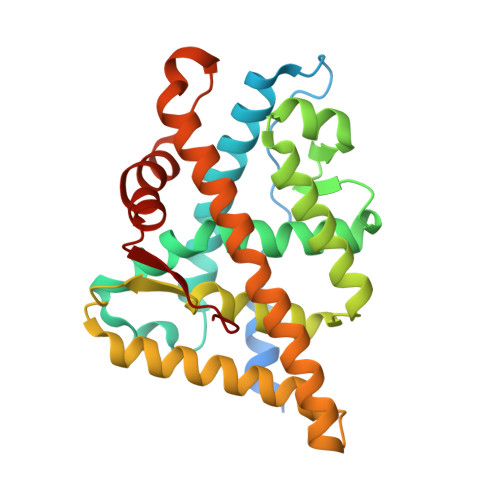
8E1A - PubMed Abstract:
The mutation-driven transformation of clinical anti-androgen drugs into agonists of the human androgen receptor (AR) represents a major challenge for the treatment of prostate cancer patients. To address this challenge, we have developed a novel class of inhibitors targeting the DNA-binding domain (DBD) of the receptor, which is distanced from the androgen binding site (ABS) targeted by all conventional anti-AR drugs and prone to resistant mutations. While many members of the developed 4-(4-phenylthiazol-2-yl)morpholine series of AR-DBD inhibitors demonstrated the effective suppression of wild-type AR, a few represented by 4-(4-(3-fluoro-2-methoxyphenyl)thiazol-2-yl)morpholine (VPC14368) exhibited a partial agonistic effect toward the mutated T878A form of the receptor, implying their cross-interaction with the AR ABS. To study the molecular basis of the observed cross-reactivity, we co-crystallized the T878A mutated form of the AR ligand binding domain (LBD) with a bound VPC14368 molecule. Computational modelling revealed that helix 12 of AR undergoes a characteristic shift upon VPC14368 binding causing the agonistic behaviour. Based on the obtained structural data we then designed derivatives of VPC14368 to successfully eliminate the cross-reactivity towards the AR ABS, while maintaining significant anti-AR DBD potency.
- Vancouver Prostate Centre, University of British Columbia, 2660 Oak Street, Vancouver, BC V6H 3Z6, Canada.
Organizational Affiliation: