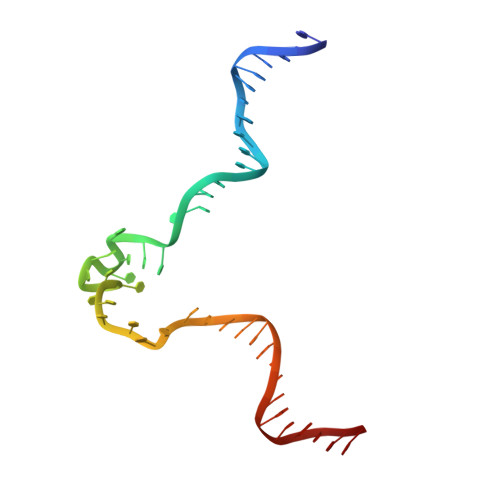
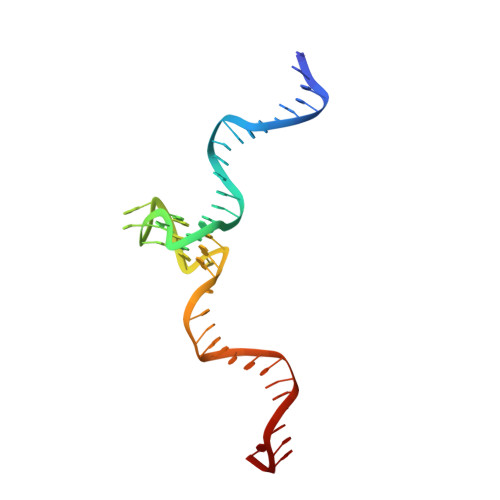
Structure of a 28.5 kDa duplex-embedded G-quadruplex system resolved to 7.4 angstrom resolution with cryo-EM.
Monsen, R.C., Chua, E.Y.D., Hopkins, J.B., Chaires, J.B., Trent, J.O.(2023) Nucleic Acids Res 51: 1943-1959
- PubMed: 36715343
- DOI: https://doi.org/10.1093/nar/gkad014
- Primary Citation of Related Structures:
8DUT - PubMed Abstract:
Genomic regions with high guanine content can fold into non-B form DNA four-stranded structures known as G-quadruplexes (G4s). Extensive in vivo investigations have revealed that promoter G4s are transcriptional regulators. Little structural information exists for these G4s embedded within duplexes, their presumed genomic environment. Here, we report the 7.4 Å resolution structure and dynamics of a 28.5 kDa duplex-G4-duplex (DGD) model system using cryo-EM, molecular dynamics, and small-angle X-ray scattering (SAXS) studies. The DGD cryo-EM refined model features a 53° bend induced by a stacked duplex-G4 interaction at the 5' G-tetrad interface with a persistently unstacked 3' duplex. The surrogate complement poly dT loop preferably stacks onto the 3' G-tetrad interface resulting in occlusion of both 5' and 3' tetrad interfaces. Structural analysis shows that the DGD model is quantifiably more druggable than the monomeric G4 structure alone and represents a new structural drug target. Our results illustrate how the integration of cryo-EM, MD, and SAXS can reveal complementary detailed static and dynamic structural information on DNA G4 systems.
- UofL Health Brown Cancer Center, University of Louisville, Louisville, KY 40202, USA.
Organizational Affiliation: