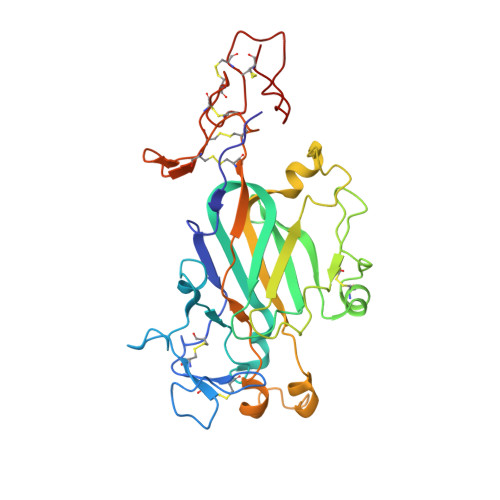
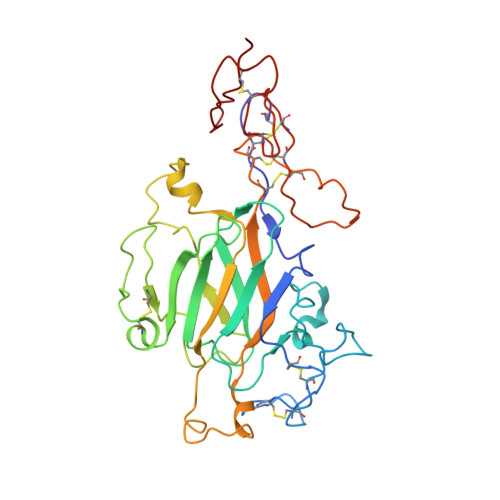
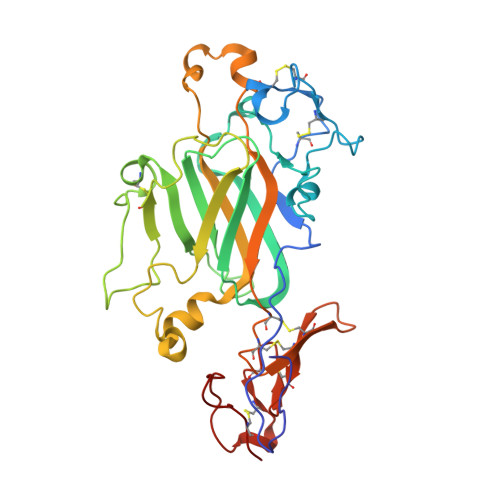
Cryo-EM reveals the molecular basis oflaminin polymerization and LN-lamininopathies.
Kulczyk, A.W., McKee, K.K., Zhang, X., Bizukojc, I., Yu, Y.Q., Yurchenco, P.D.(2023) Nat Commun 14: 317-317
- PubMed: 36658135
- DOI: https://doi.org/10.1038/s41467-023-36077-z
- Primary Citation of Related Structures:
8DMK - PubMed Abstract:
Laminin polymerization is the major step in basement membranes assembly. Its failures cause laminin N-terminal domain lamininopathies including Pierson syndrome. We have employed cryo-electron microscopy to determine a 3.7 Å structure of the trimeric laminin polymer node containing α1, β1 and γ1 subunits. The structure reveals the molecular basis of calcium-dependent formation of laminin lattice, and provides insights into polymerization defects manifesting in human disease.
- Institute for Quantitative Biomedicine, Department of Biochemistry and Microbiology, Rutgers University, Piscataway, NJ, 08854, USA. arek.kulczyk@rutgers.edu.
Organizational Affiliation: