Insight into the conformational dynamics of cytochrome P450 CYP102A1 enzyme using Cryo-EM
Su, M., Xu, H.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Bifunctional cytochrome P450/NADPH--P450 reductase | 1,037 | Priestia megaterium NBRC 15308 = ATCC 14581 | Mutation(s): 0 EC: 1.14.14.1 (UniProt), 1.6.2.4 (UniProt) | 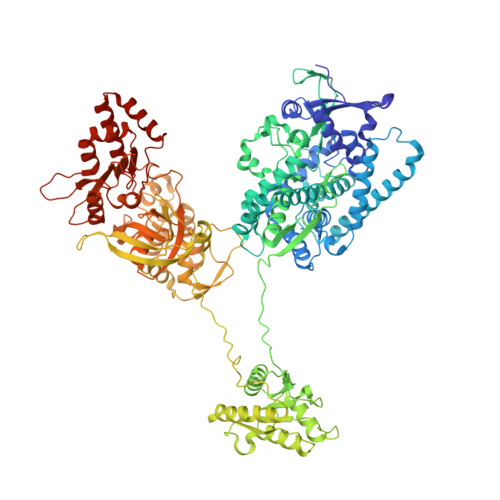 | |
UniProt | |||||
Find proteins for P14779 (Priestia megaterium (strain ATCC 14581 / DSM 32 / CCUG 1817 / JCM 2506 / NBRC 15308 / NCIMB 9376 / NCTC 10342 / NRRL B-14308 / VKM B-512 / Ford 19)) Explore P14779 Go to UniProtKB: P14779 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P14779 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD (Subject of Investigation/LOI) Query on FAD | F [auth A], O [auth B] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| HEM (Subject of Investigation/LOI) Query on HEM | C [auth A], L [auth B] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| FMN (Subject of Investigation/LOI) Query on FMN | E [auth A], N [auth B] | FLAVIN MONONUCLEOTIDE C17 H21 N4 O9 P FVTCRASFADXXNN-SCRDCRAPSA-N |  | ||
| 1C6 Query on 1C6 | D [auth A], M [auth B] | 6-methoxy-2-{[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]sulfanyl}-1H-benzimidazole C17 H19 N3 O2 S XURCIPRUUASYLR-UHFFFAOYSA-N |  | ||
| PG4 (Subject of Investigation/LOI) Query on PG4 | J [auth A], K [auth A], S [auth B], T [auth B] | TETRAETHYLENE GLYCOL C8 H18 O5 UWHCKJMYHZGTIT-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | G [auth A] H [auth A] I [auth A] P [auth B] Q [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |