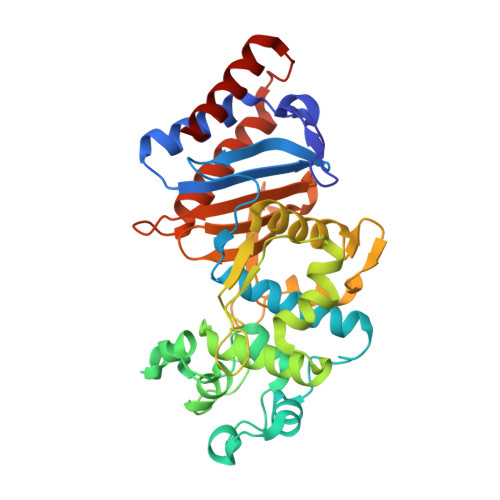
Structural and biochemical characterization of Leptospira interrogans Lsa45 reveals a penicillin-binding protein with esterase activity.
Santos, J.C., Handa, S., Fernandes, L.G.V., Bleicher, L., Gandin, C.A., de Oliveira-Neto, M., Ghosh, P., Nascimento, A.L.T.O.(2023) Process Biochem 125: 141-153
- PubMed: 36643388
- DOI: https://doi.org/10.1016/j.procbio.2022.12.010
- Primary Citation of Related Structures:
8DC1 - PubMed Abstract:
Leptospirosis is a bacterial disease that affects humans and animals and is caused by Leptospira . The recommended treatment for leptospirosis is antibiotic therapy, which should be given early in the course of the disease. Despite the use of these antibiotics, their role during the course of the disease is still not completely clear because of the lack of effective clinical trials, particularly for severe cases of the disease. Here, we present the characterization of L. interrogans Lsa45 protein by gel filtration, protein crystallography, SAXS, fluorescence and enzymatic assays. The oligomeric studies revealed that Lsa45 is monomeric in solution. The crystal structure of Lsa45 revealed the presence of two subdomains: a large α/β subdomain and a small α-helical subdomain. The large subdomain contains the amino acids Ser122, Lys125, and Tyr217, which correspond to the catalytic triad that is essential for β-lactamase or serine hydrolase activity in similar enzymes. Additionally, we also confirmed the bifunctional promiscuity of Lsa45, in hydrolyzing both the 4-nitrophenyl acetate ( p -NPA) and nitrocefin β-lactam antibiotic. Therefore, this study provides novel insights into the structure and function of enzymes from L. interrogans , which furthers our understanding of this bacterium and the development of new therapies for the prevention and treatment of leptospirosis.
- Laboratório de Desenvolvimento de Vacinas, Instituto Butantan, Avenida Vital Brasil, 1500, 05503-900, São Paulo, SP, Brazil.
Organizational Affiliation: