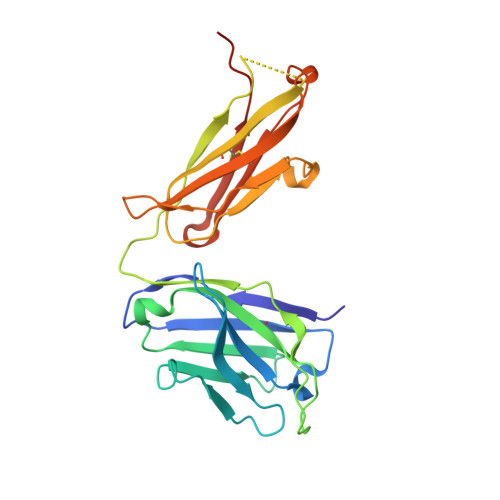
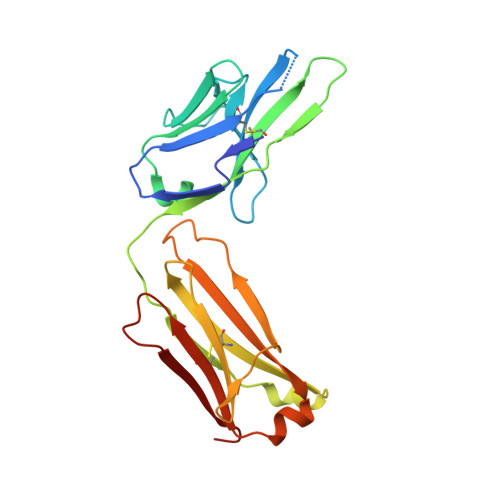
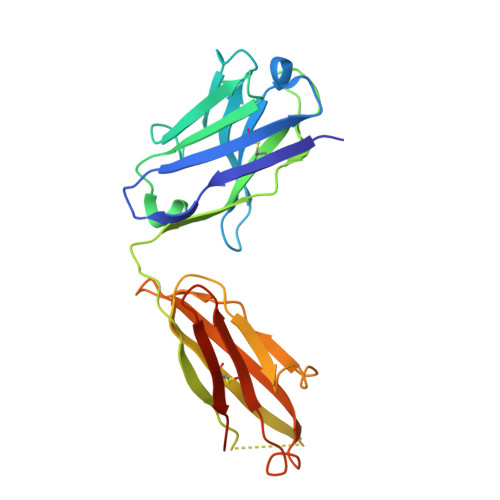
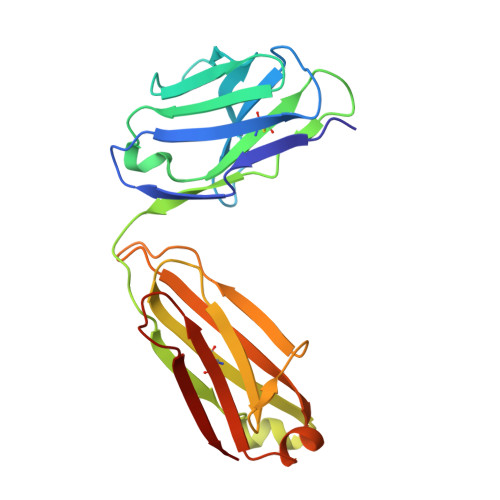
Immunotherapy-induced neutralizing antibodies disrupt allergen binding and sustain allergen tolerance in peanut allergy.
LaHood, N.A., Min, J., Keswani, T., Richardson, C.M., Amoako, K., Zhou, J., Marini-Rapoport, O., Bernard, H., Hazebrouck, S., Shreffler, W.G., Love, J.C., Pomes, A., Pedersen, L.C., Mueller, G.A., Patil, S.U.(2023) J Clin Invest 133
- PubMed: 36647835
- DOI: https://doi.org/10.1172/JCI164501
- Primary Citation of Related Structures:
8DB4 - PubMed Abstract:
In IgE-mediated food allergies, exposure to the allergen activates systemic allergic responses. Oral immunotherapy (OIT) treats food allergies through incremental increases in oral allergen exposure. However, OIT only induces sustained clinical tolerance and decreased basophil sensitivity in a subset of individuals despite increases in circulating allergen-specific IgG in all treated individuals. Therefore, we examined the allergen-specific antibodies from 2 OIT cohorts of patients with sustained and transient responses. Here, we compared antibodies from individuals with sustained or transient responses and discovered specific tolerance-associated conformational epitopes of the immunodominant allergen Ara h 2 recognized by neutralizing antibodies. First, we identified what we believe to be previously unknown conformational, intrahelical epitopes using x-ray crystallography with recombinant antibodies. We then identified epitopes only recognized in sustained tolerance. Finally, antibodies recognizing tolerance-associated epitopes effectively neutralized allergen to suppress IgE-mediated effector cell activation. Our results demonstrate the molecular basis of antibody-mediated protection in IgE-mediated food allergy, by defining how these antibodies disrupt IgE-allergen interactions to prevent allergic reactions. Our approach to studying the structural and functional basis for neutralizing antibodies demonstrates the clinical relevance of specific antibody clones in antibody-mediated tolerance. We anticipate that our findings will form the foundation for treatments of peanut allergy using neutralizing antibodies and hypoallergens.
- Food Allergy Center and Center for Immunology and Inflammatory Diseases, Massachusetts General Hospital, Boston, Massachusetts, USA.
Organizational Affiliation: