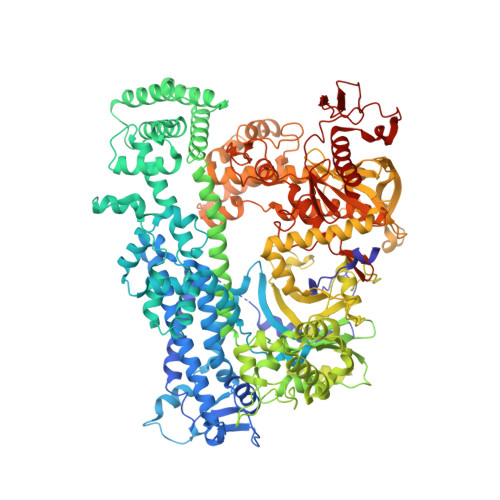
RNA targeting unleashes indiscriminate nuclease activity of CRISPR-Cas12a2.
Bravo, J.P.K., Hallmark, T., Naegle, B., Beisel, C.L., Jackson, R.N., Taylor, D.W.(2023) Nature 613: 582-587
- PubMed: 36599980
- DOI: https://doi.org/10.1038/s41586-022-05560-w
- Primary Citation of Related Structures:
8D49, 8D4A, 8D4B - PubMed Abstract:
Cas12a2 is a CRISPR-associated nuclease that performs RNA-guided, sequence-nonspecific degradation of single-stranded RNA, single-stranded DNA and double-stranded DNA following recognition of a complementary RNA target, culminating in abortive infection 1 . Here we report structures of Cas12a2 in binary, ternary and quaternary complexes to reveal a complete activation pathway. Our structures reveal that Cas12a2 is autoinhibited until binding a cognate RNA target, which exposes the RuvC active site within a large, positively charged cleft. Double-stranded DNA substrates are captured through duplex distortion and local melting, stabilized by pairs of 'aromatic clamp' residues that are crucial for double-stranded DNA degradation and in vivo immune system function. Our work provides a structural basis for this mechanism of abortive infection to achieve population-level immunity, which can be leveraged to create rational mutants that degrade a spectrum of collateral substrates.
- Department of Molecular Biosciences, University of Texas at Austin, Austin, TX, USA.
Organizational Affiliation: