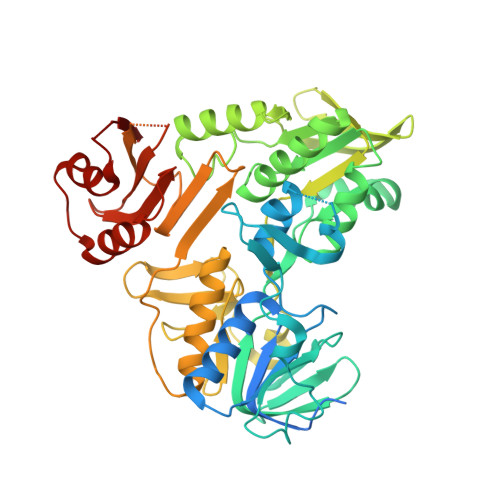
Chemical screening by time-resolved X-ray scattering to discover allosteric probes.
Brosey, C.A., Link, T.M., Shen, R., Moiani, D., Burnett, K., Hura, G.L., Jones, D.E., Tainer, J.A.(2024) Nat Chem Biol
- PubMed: 38671223
- DOI: https://doi.org/10.1038/s41589-024-01609-1
- Primary Citation of Related Structures:
8D3E, 8D3G, 8D3H, 8D3I, 8D3J, 8D3K, 8D3N, 8D3O - PubMed Abstract:
Drug discovery relies on efficient identification of small-molecule leads and their interactions with macromolecular targets. However, understanding how chemotypes impact mechanistically important conformational states often remains secondary among high-throughput discovery methods. Here, we present a conformational discovery pipeline integrating time-resolved, high-throughput small-angle X-ray scattering (TR-HT-SAXS) and classic fragment screening applied to allosteric states of the mitochondrial import oxidoreductase apoptosis-inducing factor (AIF). By monitoring oxidized and X-ray-reduced AIF states, TR-HT-SAXS leverages structure and kinetics to generate a multidimensional screening dataset that identifies fragment chemotypes allosterically stimulating AIF dimerization. Fragment-induced dimerization rates, quantified with time-resolved SAXS similarity analysis (k VR ), capture structure-activity relationships (SAR) across the top-ranked 4-aminoquinoline chemotype. Crystallized AIF-aminoquinoline complexes validate TR-SAXS-guided SAR, supporting this conformational chemotype for optimization. AIF-aminoquinoline structures and mutational analysis reveal active site F482 as an underappreciated allosteric stabilizer of AIF dimerization. This conformational discovery pipeline illustrates TR-HT-SAXS as an effective technology for targeting chemical leads to important macromolecular states.
- Department of Molecular and Cellular Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA. cabrosey@mdanderson.org.
Organizational Affiliation: