Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy
Liu, W., Zhang, X.K., Gong, D.Y., Dai, X.H., Sharma, A., Zhang, T.H., Rey, F., Zhou, Z.H.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ankyrin repeat family A protein 2,Envelope E protein | A, E [auth B], I [auth C] | 639 | Zika virus | Mutation(s): 0 Gene Names: ANKRA2, ANKRA | 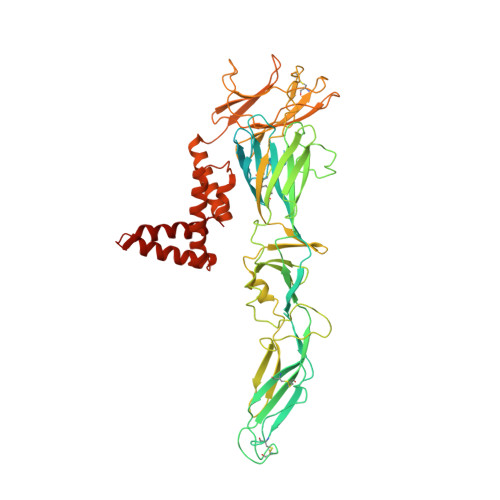 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for A0A024B7W1 (Zika virus (isolate ZIKV/Human/French Polynesia/10087PF/2013)) Explore A0A024B7W1 Go to UniProtKB: A0A024B7W1 | |||||
Find proteins for Q9H9E1 (Homo sapiens) Explore Q9H9E1 Go to UniProtKB: Q9H9E1 | |||||
PHAROS: Q9H9E1 GTEx: ENSG00000164331 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | A0A024B7W1Q9H9E1 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | Go to GlyGen: Q9H9E1-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Membrane M protein | B [auth D], F [auth E], J [auth F] | 3,423 | Zika virus | Mutation(s): 0 | 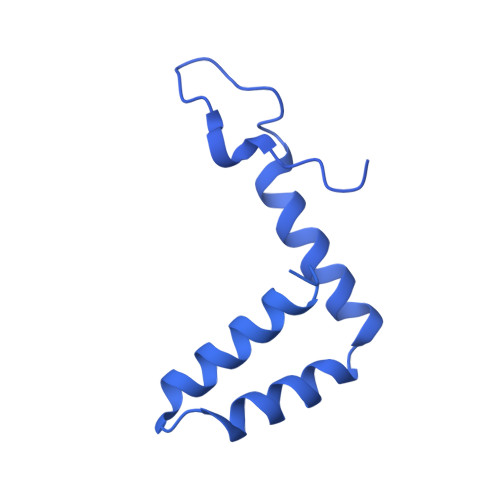 |
UniProt | |||||
Find proteins for A0A024B7W1 (Zika virus (isolate ZIKV/Human/French Polynesia/10087PF/2013)) Explore A0A024B7W1 Go to UniProtKB: A0A024B7W1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A024B7W1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| A11 heavy chain | C [auth H], G [auth h] | 133 | Homo sapiens | Mutation(s): 0 | 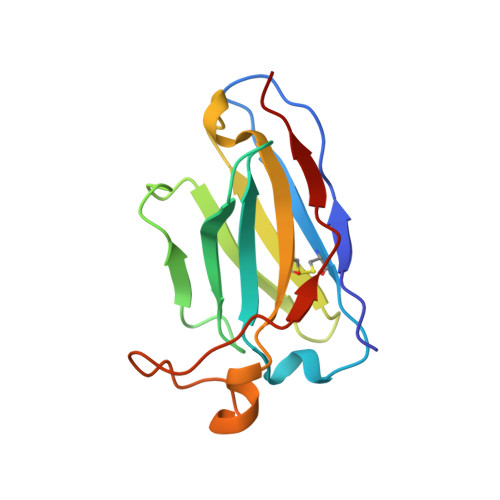 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| A11 light chain | D [auth L], H [auth l] | 111 | Homo sapiens | Mutation(s): 0 | 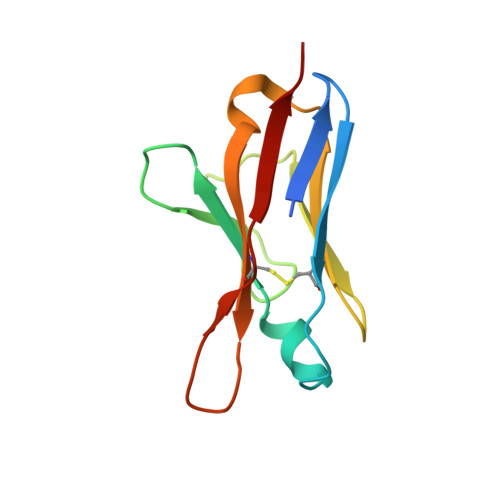 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | K [auth G], L [auth I] | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G32152BH GlyCosmos: G32152BH GlyGen: G32152BH | |||||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of Dental and Craniofacial Research (NIH/NIDCR) | United States | DE028583, DE025567 |
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | 1S10RR23057, 1S10OD018111, and 1U24GM116792 |
| National Science Foundation (NSF, United States) | United States | DBI-1338135 and DMR-1548924 |