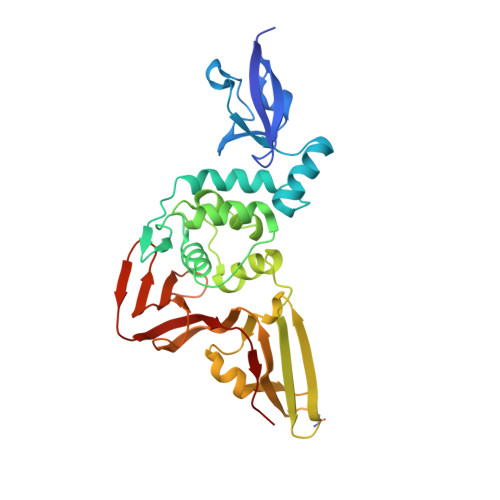
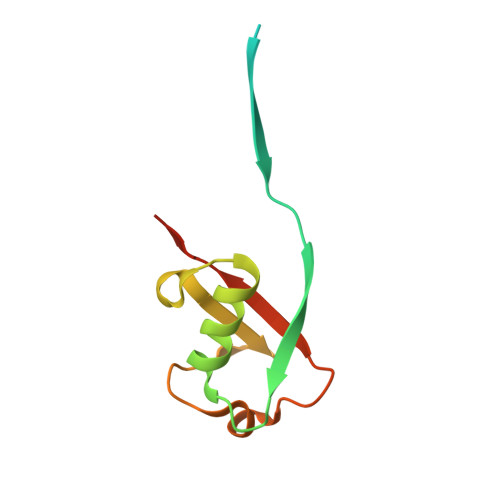
Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site.
van Vliet, V.J.E., Huynh, N., Pala, J., Patel, A., Singer, A., Slater, C., Chung, J., van Huizen, M., Teyra, J., Miersch, S., Luu, G.K., Ye, W., Sharma, N., Ganaie, S.S., Russell, R., Chen, C., Maynard, M., Amarasinghe, G.K., Mark, B.L., Kikkert, M., Sidhu, S.S.(2022) PLoS Pathog 18: e1011065-e1011065
- PubMed: 36548304
- DOI: https://doi.org/10.1371/journal.ppat.1011065
- Primary Citation of Related Structures:
8CX9 - PubMed Abstract:
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has made it clear that combating coronavirus outbreaks benefits from a combination of vaccines and therapeutics. A promising drug target common to all coronaviruses-including SARS-CoV, MERS-CoV, and SARS-CoV-2-is the papain-like protease (PLpro). PLpro cleaves part of the viral replicase polyproteins into non-structural protein subunits, which are essential to the viral replication cycle. Additionally, PLpro can cleave both ubiquitin and the ubiquitin-like protein ISG15 from host cell substrates as a mechanism to evade innate immune responses during infection. These roles make PLpro an attractive antiviral drug target. Here we demonstrate that ubiquitin variants (UbVs) can be selected from a phage-displayed library and used to specifically and potently block SARS-CoV-2 PLpro activity. A crystal structure of SARS-CoV-2 PLpro in complex with a representative UbV reveals a dimeric UbV bound to PLpro at a site distal to the catalytic site. Yet, the UbV inhibits the essential cleavage activities of the protease in vitro and in cells, and it reduces viral replication in cell culture by almost five orders of magnitude.
- Department of Medical Microbiology, Leiden University Center of Infectious Diseases (LU-CID), Leiden University Medical Center, Leiden, South Holland, The Netherlands.
Organizational Affiliation: