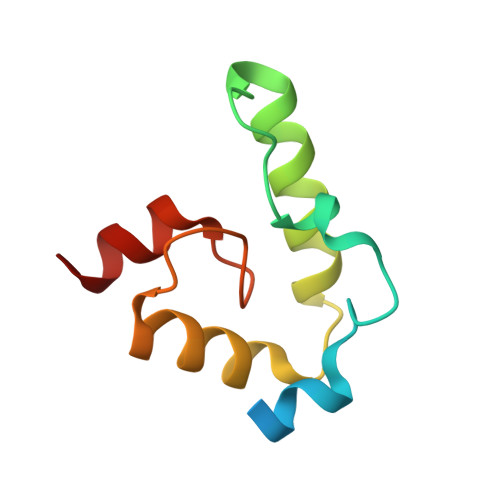
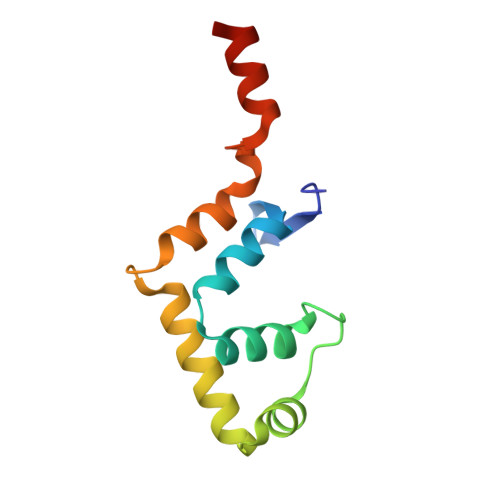
Structural basis of DNA binding by the WhiB-like transcription factor WhiB3 in Mycobacterium tuberculosis.
Wan, T., Horova, M., Khetrapal, V., Li, S., Jones, C., Schacht, A., Sun, X., Zhang, L.(2023) J Biological Chem 299: 104777-104777
- PubMed: 37142222
- DOI: https://doi.org/10.1016/j.jbc.2023.104777
- Primary Citation of Related Structures:
8CWR, 8CWT, 8CYF - PubMed Abstract:
Mycobacterium tuberculosis (Mtb) WhiB3 is an iron-sulfur cluster-containing transcription factor belonging to a subclass of the WhiB-Like (Wbl) family that is widely distributed in the phylum Actinobacteria. WhiB3 plays a crucial role in the survival and pathogenesis of Mtb. It binds to the conserved region 4 of the principal sigma factor (σ A 4 ) in the RNA polymerase holoenzyme to regulate gene expression like other known Wbl proteins in Mtb. However, the structural basis of how WhiB3 coordinates with σ A 4 to bind DNA and regulate transcription is unclear. Here we determined crystal structures of the WhiB3:σ A 4 complex without and with DNA at 1.5 Å and 2.45 Å, respectively, to elucidate how WhiB3 interacts with DNA to regulate gene expression. These structures reveal that the WhiB3:σ A 4 complex shares a molecular interface similar to other structurally characterized Wbl proteins and also possesses a subclass-specific Arg-rich DNA-binding motif. We demonstrate that this newly defined Arg-rich motif is required for WhiB3 binding to DNA in vitro and transcriptional regulation in Mycobacterium smegmatis. Together, our study provides empirical evidence of how WhiB3 regulates gene expression in Mtb by partnering with σ A 4 and engaging with DNA via the subclass-specific structural motif, distinct from the modes of DNA interaction by WhiB1 and WhiB7.
- Department of Biochemistry, University of Nebraska-Lincoln, Lincoln, Nebraska, USA.
Organizational Affiliation: