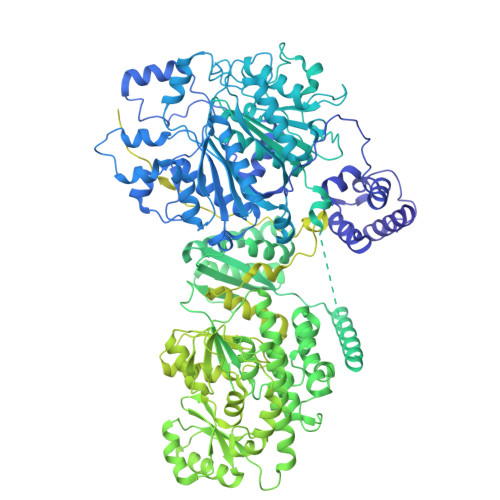
Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Kim, S.K., Dickinson, M.S., Finer-Moore, J., Guan, Z., Kaake, R.M., Echeverria, I., Chen, J., Pulido, E.H., Sali, A., Krogan, N.J., Rosenberg, O.S., Stroud, R.M.(2023) Nat Struct Mol Biol 30: 296-308
- PubMed: 36782050
- DOI: https://doi.org/10.1038/s41594-022-00918-0
- Primary Citation of Related Structures:
7UK4, 8CUY, 8CUZ, 8CV0, 8CV1 - PubMed Abstract:
The mycolic acid layer of the Mycobacterium tuberculosis cell wall is essential for viability and virulence, and the enzymes responsible for its synthesis are targets for antimycobacterial drug development. Polyketide synthase 13 (Pks13) is a module encoding several enzymatic and transport functions that carries out the condensation of two different long-chain fatty acids to produce mycolic acids. We determined structures by cryogenic-electron microscopy of dimeric multi-enzyme Pks13 purified from mycobacteria under normal growth conditions, captured with native substrates. Structures define the ketosynthase (KS), linker and acyl transferase (AT) domains at 1.8 Å resolution and two alternative locations of the N-terminal acyl carrier protein. These structures suggest intermediate states on the pathway for substrate delivery to the KS domain. Other domains, visible at lower resolution, are flexible relative to the KS-AT core. The chemical structures of three bound endogenous long-chain fatty acid substrates were determined by electrospray ionization mass spectrometry.
- Department of Biochemistry and Biophysics, University of California San Francisco, San Francisco, CA, USA.
Organizational Affiliation: