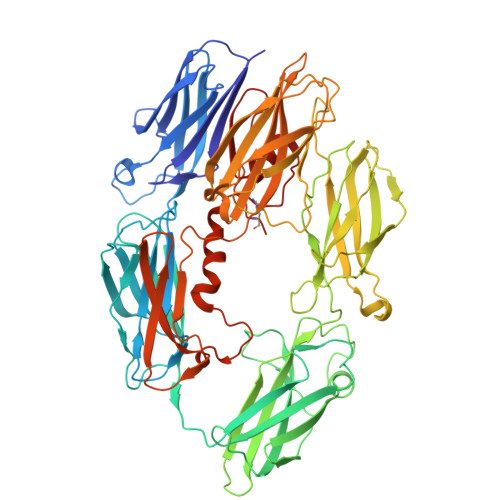
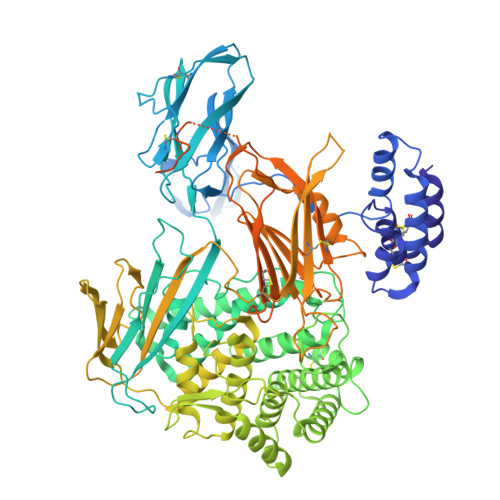
Inhibition of cleavage of human complement component C5 and the R885H C5 variant by two distinct high affinity anti-C5 nanobodies.
Struijf, E.M., De la O Becerra, K.I., Ruyken, M., de Haas, C.J.C., van Oosterom, F., Siere, D.Y., van Keulen, J.E., Heesterbeek, D.A.C., Dolk, E., Heukers, R., Bardoel, B.W., Gros, P., Rooijakkers, S.H.M.(2023) J Biological Chem 299: 104956-104956
- PubMed: 37356719
- DOI: https://doi.org/10.1016/j.jbc.2023.104956
- Primary Citation of Related Structures:
8CML - PubMed Abstract:
The human complement system plays a crucial role in immune defense. However, its erroneous activation contributes to many serious inflammatory diseases. Since most unwanted complement effector functions result from C5 cleavage into C5a and C5b, development of C5 inhibitors, such as clinically approved monoclonal antibody eculizumab, are of great interest. Here, we developed and characterized two anti-C5 nanobodies, UNbC5-1 and UNbC5-2. Using surface plasmon resonance, we determined a binding affinity of 119.9 pM for UNbC5-1 and 7.7 pM for UNbC5-2. Competition experiments determined that the two nanobodies recognize distinct epitopes on C5. Both nanobodies efficiently interfered with C5 cleavage in a human serum environment, as they prevented red blood cell lysis via membrane attack complexes (C5b-9) and the formation of chemoattractant C5a. The cryo-EM structure of UNbC5-1 and UNbC5-2 in complex with C5 (3.6 Å resolution) revealed that the binding interfaces of UNbC5-1 and UNbC5-2 overlap with known complement inhibitors eculizumab and RaCI3, respectively. UNbC5-1 binds to the MG7 domain of C5, facilitated by a hydrophobic core and polar interactions, and UNbC5-2 interacts with the C5d domain mostly by salt bridges and hydrogen bonds. Interestingly, UNbC5-1 potently binds and inhibits C5 R885H, a genetic variant of C5 that is not recognized by eculizumab. Altogether, we identified and characterized two different, high affinity nanobodies against human C5. Both nanobodies could serve as diagnostic and/or research tools to detect C5 or inhibit C5 cleavage. Furthermore, the residues targeted by UNbC5-1 hold important information for therapeutic inhibition of different polymorphic variants of C5.
- Department Medical Microbiology, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Organizational Affiliation: