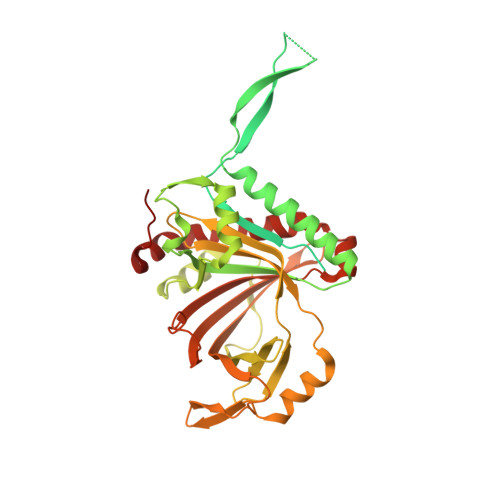
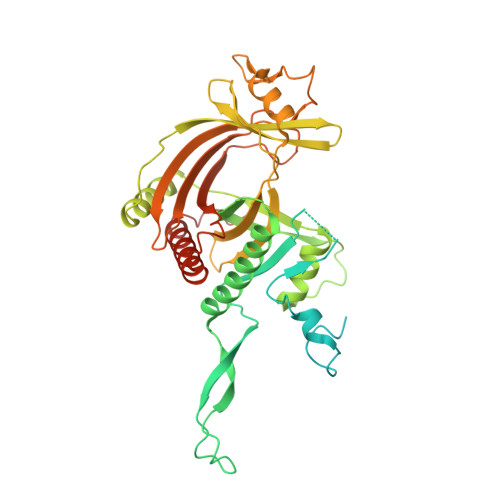
Structural basis for guide RNA selection by the RESC1-RESC2 complex.
Dolce, L.G., Nesterenko, Y., Walther, L., Weis, F., Kowalinski, E.(2023) Nucleic Acids Res 51: 4602-4612
- PubMed: 36999600
- DOI: https://doi.org/10.1093/nar/gkad217
- Primary Citation of Related Structures:
8CDP - PubMed Abstract:
Kinetoplastid parasites, such as trypanosomes or leishmania, rely on RNA-templated RNA editing to mature mitochondrial cryptic pre-mRNAs into functional protein-coding transcripts. Processive pan-editing of multiple editing blocks within a single transcript is dependent on the 20-subunit RNA editing substrate binding complex (RESC) that serves as a platform to orchestrate the interactions between pre-mRNA, guide RNAs (gRNAs), the catalytic RNA editing complex (RECC), and a set of RNA helicases. Due to the lack of molecular structures and biochemical studies with purified components, neither the spacio-temporal interplay of these factors nor the selection mechanism for the different RNA components is understood. Here we report the cryo-EM structure of Trypanosoma brucei RESC1-RESC2, a central hub module of the RESC complex. The structure reveals that RESC1 and RESC2 form an obligatory domain-swapped dimer. Although the tertiary structures of both subunits closely resemble each other, only RESC2 selectively binds 5'-triphosphate-nucleosides, a defining characteristic of gRNAs. We therefore propose RESC2 as the protective 5'-end binding site for gRNAs within the RESC complex. Overall, our structure provides a starting point for the study of the assembly and function of larger RNA-bound kinetoplast RNA editing modules and might aid in the design of anti-parasite drugs.
- EMBL Grenoble, 71 Avenue des Martyrs, 38042 Grenoble, France.
Organizational Affiliation: