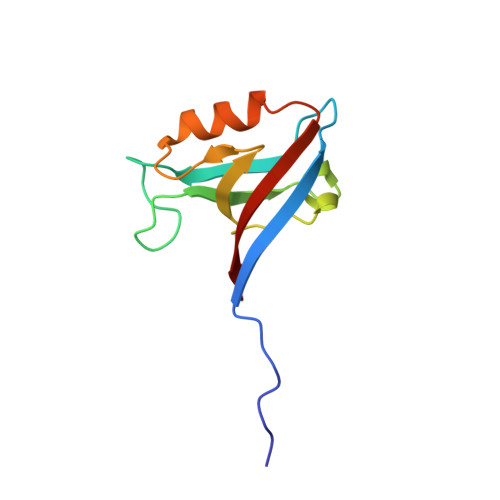
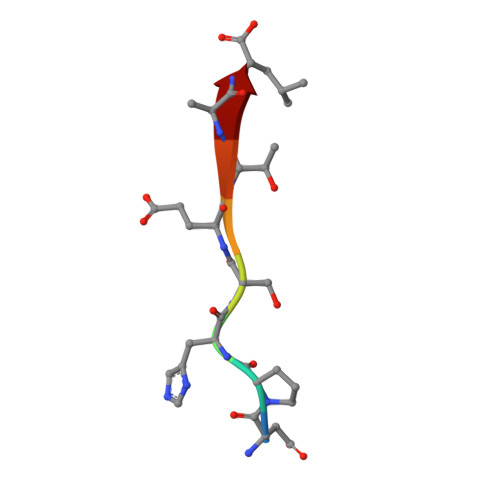
Membrane recruitment of the polarity protein Scribble by the cell adhesion receptor TMIGD1.
Thuring, E.M., Hartmann, C., Maddumage, J.C., Javorsky, A., Michels, B.E., Gerke, V., Banks, L., Humbert, P.O., Kvansakul, M., Ebnet, K.(2023) Commun Biol 6: 702-702
- PubMed: 37430142
- DOI: https://doi.org/10.1038/s42003-023-05088-3
- Primary Citation of Related Structures:
8CD3 - PubMed Abstract:
Scribble (Scrib) is a multidomain polarity protein and member of the leucine-rich repeat and PDZ domain (LAP) protein family. A loss of Scrib expression is associated with disturbed apical-basal polarity and tumor formation. The tumor-suppressive activity of Scrib correlates with its membrane localization. Despite the identification of numerous Scrib-interacting proteins, the mechanisms regulating its membrane recruitment are not fully understood. Here, we identify the cell adhesion receptor TMIGD1 as a membrane anchor of Scrib. TMIGD1 directly interacts with Scrib through a PDZ domain-mediated interaction and recruits Scrib to the lateral membrane domain in epithelial cells. We characterize the association of TMIGD1 with each Scrib PDZ domain and describe the crystal structure of the TMIGD1 C-terminal peptide complexed with PDZ domain 1 of Scrib. Our findings describe a mechanism of Scrib membrane localization and contribute to the understanding of the tumor-suppressive activity of Scrib.
- Institute-associated Research Group "Cell adhesion and cell polarity", Institute of Medical Biochemistry, ZMBE, University of Münster, Münster, Germany.
Organizational Affiliation: