Structure of IgE bound to the ectodomain of FceRIa
Andersen, G.R., Jensen, R.K.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Immunoglobulin heavy constant epsilon | A [auth H], D [auth X] | 426 | Homo sapiens | Mutation(s): 0 Gene Names: IGHE | 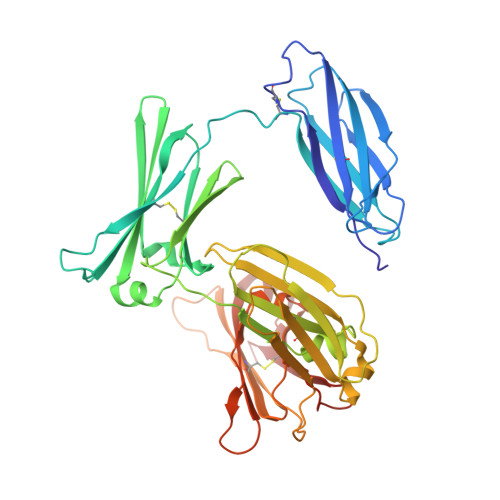 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P01854 (Homo sapiens) Explore P01854 Go to UniProtKB: P01854 | |||||
PHAROS: P01854 GTEx: ENSG00000211891 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01854 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | Go to GlyGen: P01854-2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Immunoglobulin kappa constant | B [auth L], E [auth Y] | 107 | Homo sapiens | Mutation(s): 0 Gene Names: IGKC | 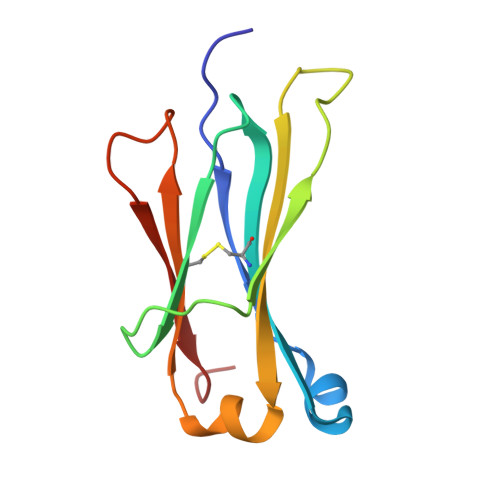 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P01834 (Homo sapiens) Explore P01834 Go to UniProtKB: P01834 | |||||
PHAROS: P01834 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01834 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| High affinity immunoglobulin epsilon receptor subunit alpha | C [auth R] | 171 | Homo sapiens | Mutation(s): 0 Gene Names: FCER1A, FCE1A | 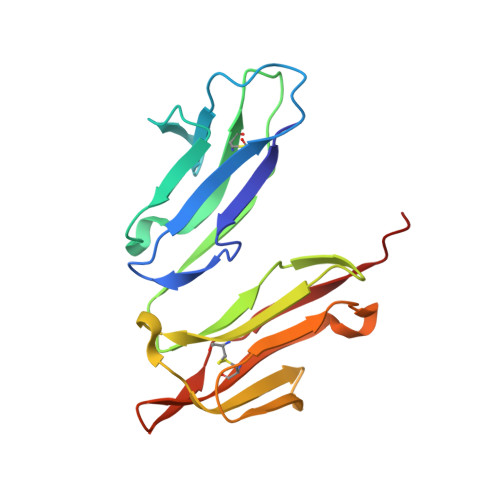 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P12319 (Homo sapiens) Explore P12319 Go to UniProtKB: P12319 | |||||
PHAROS: P12319 GTEx: ENSG00000179639 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P12319 | ||||
Glycosylation | |||||
| Glycosylation Sites: 7 | Go to GlyGen: P12319-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | F [auth A], L [auth G] | 5 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G22768VO GlyCosmos: G22768VO GlyGen: G22768VO | |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | G [auth B], H [auth C] | 5 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G21381MC GlyCosmos: G21381MC GlyGen: G21381MC | |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | M [auth R], N [auth R] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Novo Nordisk Foundation | Denmark | NNF18OC0052105 |
| Lundbeckfonden | Denmark | R155-2015-2666 |
| Danish Council for Independent Research | Denmark | 0135-00061B |