streptavidin mutant S112I with an iridium catalyst for CH activation
Igareta, N.V., Ward, T.R.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Streptavidin | 123 | Streptomyces avidinii | Mutation(s): 0 | 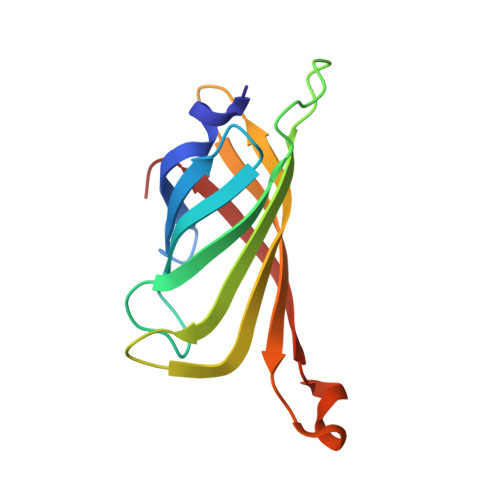 | |
UniProt | |||||
Find proteins for P22629 (Streptomyces avidinii) Explore P22629 Go to UniProtKB: P22629 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P22629 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NOF (Subject of Investigation/LOI) Query on NOF | B [auth A] | tert-butyl 7'-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]-1-chloranyl-2,3,4,5,6-pentamethyl-spiro[1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane-1,2'-3-aza-1-azonia-2$l^{8}-iridatricyclo[6.3.1.0^{4,12}]dodeca-1(11),4,6,8(12),9-pentaene]-3'-carboxylate C34 H45 Cl Ir N5 O4 S NIFXELQADIYDEE-HVXVAELJSA-L |  | ||
| SO4 Query on SO4 | C [auth A], D [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 57.657 | α = 90 |
| b = 57.657 | β = 90 |
| c = 183.626 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| Coot | model building |
| PHASER | phasing |
| Aimless | data scaling |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |