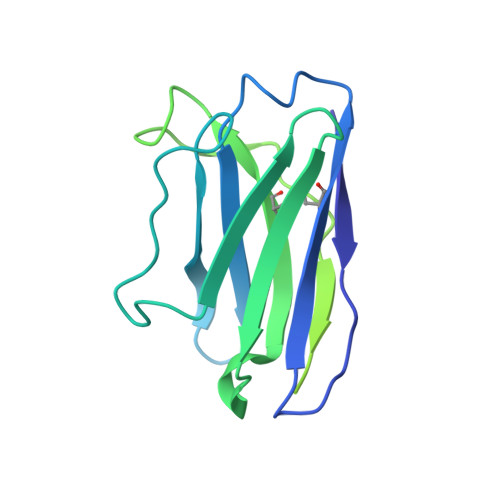
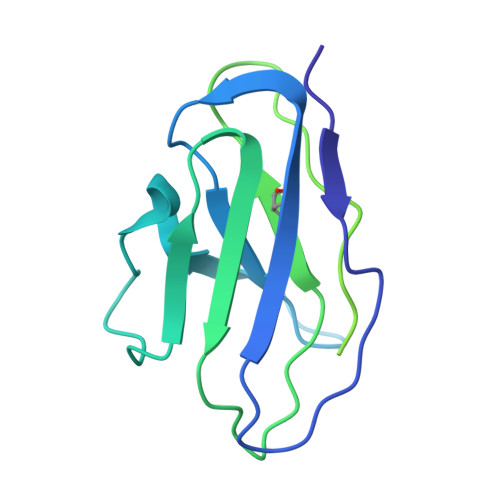
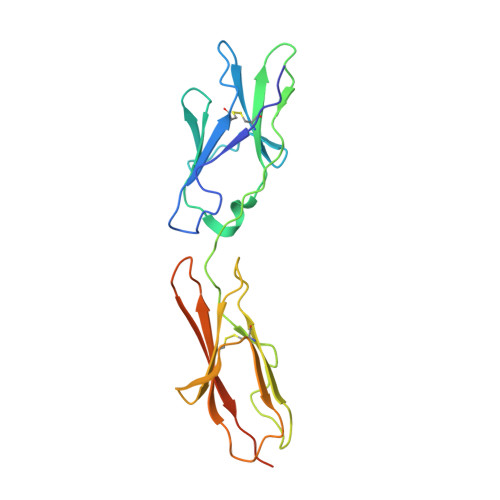
Structural insights into epitope-paratope interactions of a monoclonal antibody targeting CEACAM5-expressing tumors.
Kumar, A., Duffieux, F., Gagnaire, M., Rapisarda, C., Bertrand, T., Rak, A.(2024) Nat Commun 15: 9377-9377
- PubMed: 39477960
- DOI: https://doi.org/10.1038/s41467-024-53746-9
- Primary Citation of Related Structures:
8BW0 - PubMed Abstract:
Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) are overexpressed in some tumor types. The antibody-drug conjugate tusamitamab ravtansine specifically recognizes the A3-B3 domains of human CEACAM5 (hCEACAM5). To understand this specificity, here we map the epitope-paratope interface between the A3-B3 domains of hCEACAM5 (hCEACAM5 A3-B3 ) and the antigen-binding fragment of tusamitamab (tusa Fab). We use hydrogen/deuterium exchange mass spectrometry to identify the tusa Fab paratope, which involves heavy chain (HC) residues 101-109 and light chain residues 48-54 and 88-104. Using surface plasmon resonance, we demonstrate that alanine variants of HC residues 96-108 abolish binding to hCEACAM5, suggesting that these residues are critical for tusa-Fab-antigen complex formation. The cryogenic electron microscopy structure of the hCEACAM5 A3-B3 - tusa Fab complex (3.11 Å overall resolution) reveals a discontinuous epitope involving residues in the A3-B3 domains and an N-linked mannose at residue Asn612. Conformational constraints on the epitope-paratope interface enable tusamitamab to target hCEACAM5 A3-B3 and distinguish CEACAM5 from other CEACAMs.
- Integrated Drug Discovery, Sanofi R&D, Paris, France.
Organizational Affiliation: