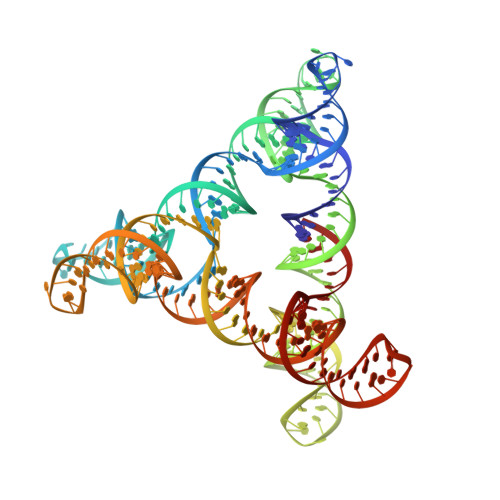
An RNA Paranemic Crossover Triangle as A 3D Module for Cotranscriptional Nanoassembly.
Sampedro Vallina, N., McRae, E.K.S., Geary, C., Andersen, E.S.(2023) Small 19: e2204651-e2204651
- PubMed: 36526605
- DOI: https://doi.org/10.1002/smll.202204651
- Primary Citation of Related Structures:
8BTZ, 8BU8 - PubMed Abstract:
RNA nanotechnology takes advantage of structural modularity to build self-assembling nano-architectures with applications in medicine and synthetic biology. The use of paranemic motifs, that form without unfolding existing secondary structure, allows for the creation of RNA nanostructures that are compatible with cotranscriptional folding in vitro and in vivo. In previous work, kissing-loop (KL) motifs have been widely used to design RNA nanostructures that fold cotranscriptionally. However, the paranemic crossover (PX) motif has not yet been explored for cotranscriptional RNA origami architectures and information about the structural geometry of the motif is unknown. Here, a six base pair-wide paranemic RNA interaction that arranges double helices in a perpendicular manner is introduced, allowing for the generation of a new and versatile building block: the paranemic-crossover triangle (PXT). The PXT is self-assembled by cotranscriptional folding and characterized by cryogenic electron microscopy, revealing for the first time an RNA PX interaction in high structural detail. The PXT is used as a building block for the construction of multimers that form filaments and rings and a duplicated PXT motif is used as a building block to self-assemble cubic structures, demonstrating the PXT as a rigid self-folding domain for the development of wireframe RNA origami architectures.
- Interdisciplinary Nanoscience Center (iNANO); Gustav Wieds Vej 14, Aarhus University, Aarhus, DK-8000, Denmark.
Organizational Affiliation: