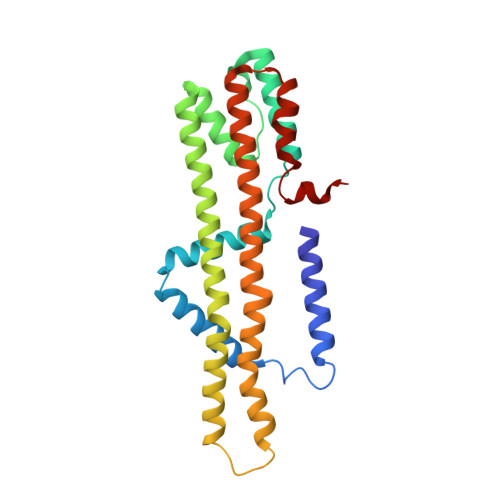
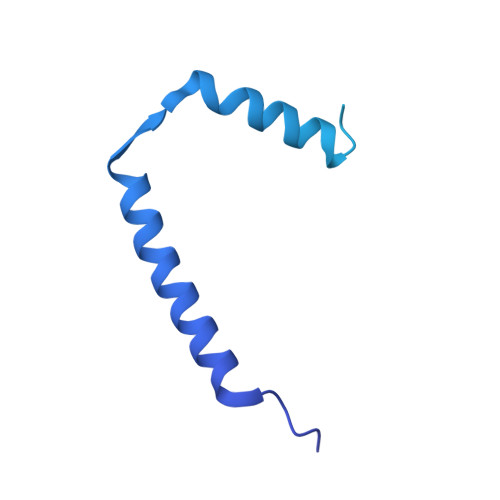
Ion selectivity and rotor coupling of the Vibrio flagellar sodium-driven stator unit.
Hu, H., Popp, P.F., Santiveri, M., Roa-Eguiara, A., Yan, Y., Martin, F.J.O., Liu, Z., Wadhwa, N., Wang, Y., Erhardt, M., Taylor, N.M.I.(2023) Nat Commun 14: 4411-4411
- PubMed: 37500658
- DOI: https://doi.org/10.1038/s41467-023-39899-z
- Primary Citation of Related Structures:
8BRD, 8BRI - PubMed Abstract:
Bacteria swim using a flagellar motor that is powered by stator units. Vibrio spp. are highly motile bacteria responsible for various human diseases, the polar flagella of which are exclusively driven by sodium-dependent stator units (PomAB). However, how ion selectivity is attained, how ion transport triggers the directional rotation of the stator unit, and how the stator unit is incorporated into the flagellar rotor remained largely unclear. Here, we have determined by cryo-electron microscopy the structure of Vibrio PomAB. The electrostatic potential map uncovers sodium binding sites, which together with functional experiments and molecular dynamics simulations, reveal a mechanism for ion translocation and selectivity. Bulky hydrophobic residues from PomA prime PomA for clockwise rotation. We propose that a dynamic helical motif in PomA regulates the distance between PomA subunit cytoplasmic domains, stator unit activation, and torque transmission. Together, our study provides mechanistic insights for understanding ion selectivity and rotor incorporation of the stator unit of the bacterial flagellum.
- Structural Biology of Molecular Machines Group, Protein Structure & Function Program, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Blegdamsvej 3B, 2200, Copenhagen, Denmark.
Organizational Affiliation: