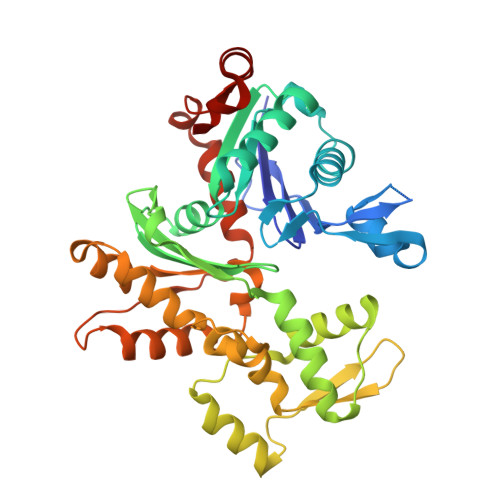
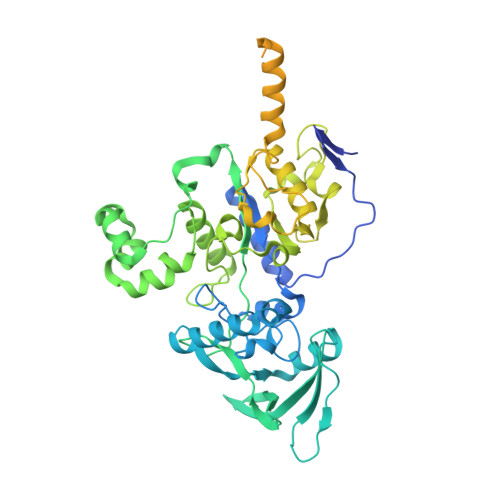
Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Teixeira-Nunes, M., Retailleau, P., Raoux-Barbot, D., Comisso, M., Missinou, A.A., Velours, C., Plancqueel, S., Ladant, D., Mechold, U., Renault, L.(2023) PLoS Pathog 19: e1011654-e1011654
- PubMed: 37747912
- DOI: https://doi.org/10.1371/journal.ppat.1011654
- Primary Citation of Related Structures:
8BJH, 8BJI, 8BJJ, 8BO1, 8BR0, 8BR1 - PubMed Abstract:
ExoY virulence factors are members of a family of bacterial nucleotidyl cyclases (NCs) that are activated by specific eukaryotic cofactors and overproduce cyclic purine and pyrimidine nucleotides in host cells. ExoYs act as actin-activated NC toxins. Here, we explore the Vibrio nigripulchritudo Multifunctional-Autoprocessing Repeats-in-ToXin (MARTX) ExoY effector domain (Vn-ExoY) as a model for ExoY-type members that interact with monomeric (G-actin) instead of filamentous (F-actin) actin. Vn-ExoY exhibits moderate binding affinity to free or profilin-bound G-actin but can capture the G-actin:profilin complex, preventing its spontaneous or VASP- or formin-mediated assembly at F-actin barbed ends in vitro. This mechanism may prolong the activated cofactor-bound state of Vn-ExoY at sites of active actin cytoskeleton remodelling. We present a series of high-resolution crystal structures of nucleotide-free, 3'-deoxy-ATP- or 3'-deoxy-CTP-bound Vn-ExoY, activated by free or profilin-bound G-actin-ATP/-ADP, revealing that the cofactor only partially stabilises the nucleotide-binding pocket (NBP) of NC toxins. Substrate binding induces a large, previously-unidentified, closure of their NBP, confining catalytically important residues and metal cofactors around the substrate, and facilitating the recruitment of two metal ions to tightly coordinate the triphosphate moiety of purine or pyrimidine nucleotide substrates. We validate critical residues for both the purinyl and pyrimidinyl cyclase activity of NC toxins in Vn-ExoY and its distantly-related ExoY from Pseudomonas aeruginosa, which specifically interacts with F-actin. The data conclusively demonstrate that NC toxins employ a similar two-metal-ion mechanism for catalysing the cyclisation of nucleotides of different sizes. These structural insights into the dynamics of the actin-binding interface of actin-activated ExoYs and the multi-step activation of all NC toxins offer new perspectives for the specific inhibition of class II bacterial NC enzymes.
- Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), Gif-sur-Yvette, France.
Organizational Affiliation: