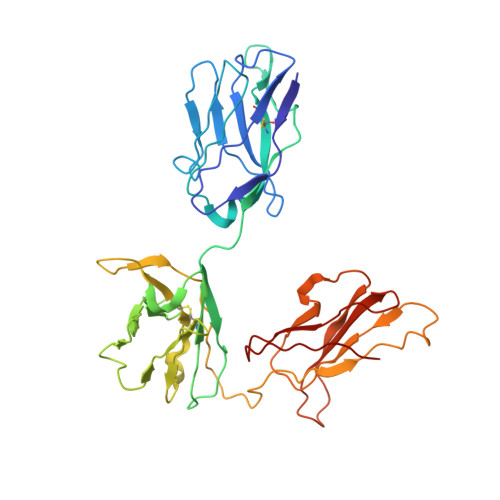
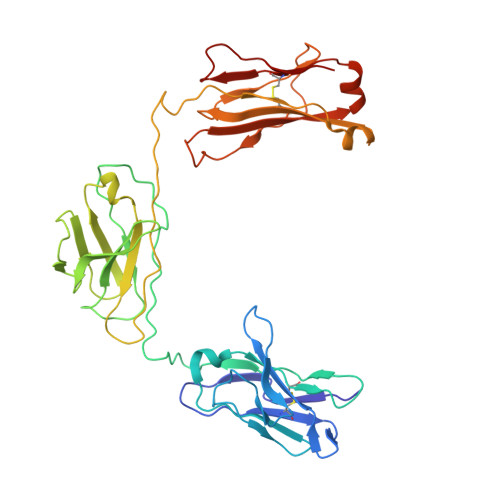
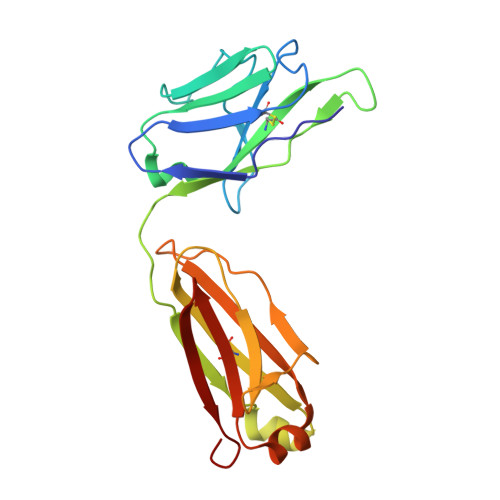
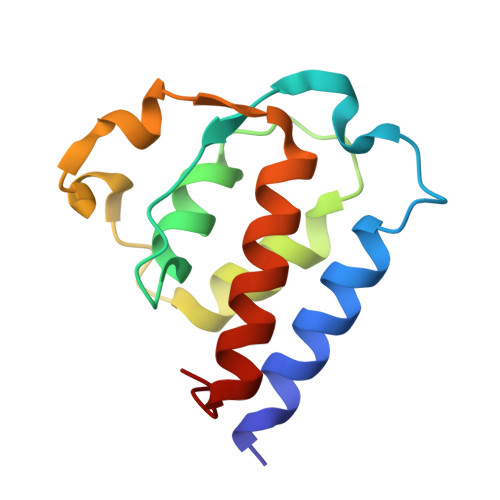
Structural insights into the bi-specific cross-over dual variable antibody architecture by cryo-EM.
Fernandez-Martinez, D., Tully, M.D., Leonard, G., Mathieu, M., Kandiah, E.(2023) Sci Rep 13: 8694-8694
- PubMed: 37248285
- DOI: https://doi.org/10.1038/s41598-023-35678-4
- Primary Citation of Related Structures:
8BLQ - PubMed Abstract:
Multi-specific antibodies (msAbs) are being developed as next generation antibody-based therapeutics. Knowledge of the three-dimensional structures, in the full antibody context, of their fragment antigen-binding (Fab) moieties with or without bound antigens is key to elucidating their therapeutic efficiency and stability. However, the flexibility of msAbs, a feature essential for their multi specificity, has hindered efforts in this direction. Cross-Over Dual Variable immunoglobulin (CODV Ig ) is a promising bispecific antibody format, designed to simultaneously target the interleukins IL4 and IL13. In this work we present the biophysical and structural characterisation of a CODV Fab :IL13 complex in the full antibody context, using cryo-electron microscopy at an overall resolution of 4.2 Å. Unlike the 1:2 stoichiometry previously observed for CODV Ig :IL4, CODV Ig :IL13 shows a 1:1 stoichiometry. As well as providing details of the IL13-CODV binding interface, including the residues involved in the epitope-paratope region, the structure of CODV Fab :IL13 also validates the use of labelling antibody as a new strategy for the single particle cryo-EM study of msAbs in complex with one, or more, antigens. This strategy reduced the inherent flexibility of the IL13 binding domain of CODV without inducing either structural changes at the epitope level or steric hindrance between the IL4 and IL13 binding regions of CODV Ig . The work presented here thus also contributes to the development of methodology for the structural study of msAbs, a promising platform for cancer immunotherapy.
- European Synchrotron Radiation Facility, 71 Avenue des Martyrs, 38042, Grenoble, France.
Organizational Affiliation: