Oxidative damage induce copper(II)-DNA binding
Lambert, M.C., Brazier, J.A., Cardin, C.J., Hall, J.P.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*(8OG)P*CP*AP*TP*GP*CP*T)-3') | 7 | synthetic construct | 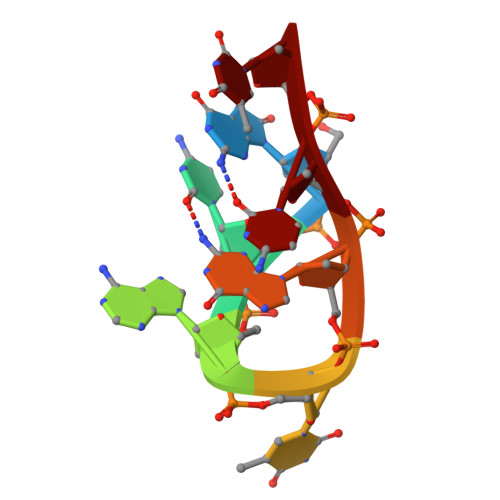 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NCO Query on NCO | C [auth A], D [auth A], E [auth A], F [auth A] | COBALT HEXAMMINE(III) Co H18 N6 DYLMFCCYOUSRTK-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 22.035 | α = 90 |
| b = 58.846 | β = 90 |
| c = 45.488 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CrysalisPro | data reduction |
| CrysalisPro | data scaling |
| PHASER | phasing |
| Coot | model building |
| PHENIX | refinement |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Engineering and Physical Sciences Research Council | United Kingdom | -- |