Crystal structure determination of a highly active UDP-glucose pyrophosphorylase from Thermocrispum agreste DSM 44070
Kumpf, A., Maier, A., Laustsen, J.U., Jeffries, C.M., Bento, I., Tischler, D.To be published.
Experimental Data Snapshot
Starting Model: other
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| UTP--glucose-1-phosphate uridylyltransferase | 299 | Thermocrispum agreste DSM 44070 | Mutation(s): 0 Gene Names: DIU77_00305 EC: 2.7.7.9 | 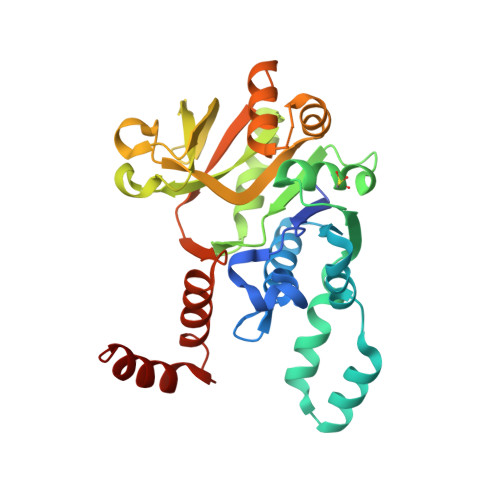 | |
UniProt | |||||
Find proteins for A0A2W4LV58 (Thermocrispum agreste) Explore A0A2W4LV58 Go to UniProtKB: A0A2W4LV58 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A2W4LV58 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| UPG (Subject of Investigation/LOI) Query on UPG | C [auth A] | URIDINE-5'-DIPHOSPHATE-GLUCOSE C15 H24 N2 O17 P2 HSCJRCZFDFQWRP-JZMIEXBBSA-N |  | ||
| PEG Query on PEG | AA [auth B], H [auth A], R [auth A], W [auth B] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | BA [auth B] CA [auth B] D [auth A] E [auth A] F [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| SCN Query on SCN | T [auth A] | THIOCYANATE ION C N S ZMZDMBWJUHKJPS-UHFFFAOYSA-M |  | ||
| MG Query on MG | V [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSX Query on CSX | A, B | L-PEPTIDE LINKING | C3 H7 N O3 S |  | CYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 48.036 | α = 90 |
| b = 78.433 | β = 93.904 |
| c = 90.289 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| European Union (EU) | European Union | 100263899 |