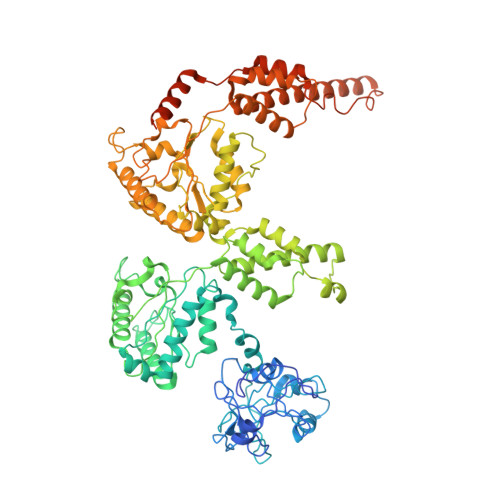
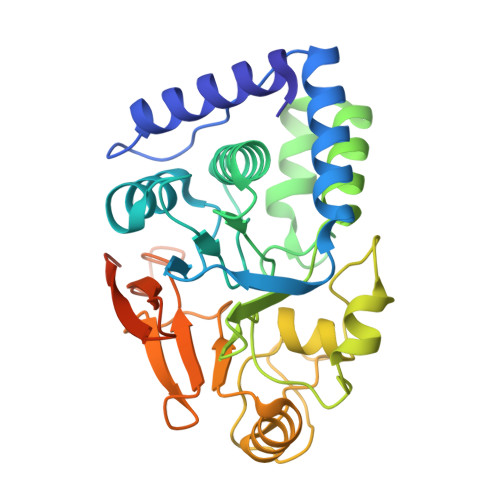
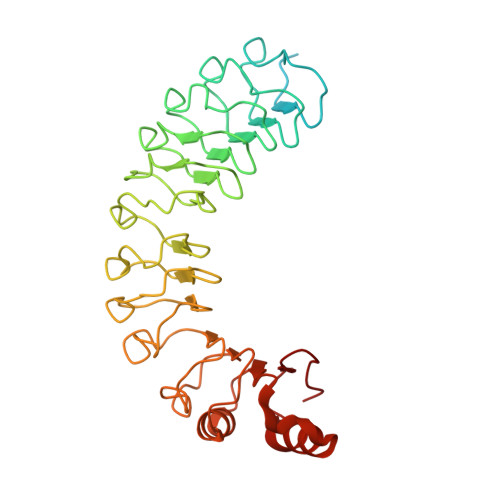
Structural basis of ubiquitin-independent PP1 complex disassembly by p97.
van den Boom, J., Marini, G., Meyer, H., Saibil, H.R.(2023) EMBO J 42: e113110-e113110
- PubMed: 37264685
- DOI: https://doi.org/10.15252/embj.2022113110
- Primary Citation of Related Structures:
8B5R - PubMed Abstract:
The AAA+-ATPase p97 (also called VCP or Cdc48) unfolds proteins and disassembles protein complexes in numerous cellular processes, but how substrate complexes are loaded onto p97 and disassembled is unclear. Here, we present cryo-EM structures of p97 in the process of disassembling a protein phosphatase-1 (PP1) complex by extracting an inhibitory subunit from PP1. We show that PP1 and its partners SDS22 and inhibitor-3 (I3) are loaded tightly onto p97, surprisingly via a direct contact of SDS22 with the p97 N-domain. Loading is assisted by the p37 adapter that bridges two adjacent p97 N-domains underneath the substrate complex. A stretch of I3 is threaded into the central channel of the spiral-shaped p97 hexamer, while other elements of I3 are still attached to PP1. Thus, our data show how p97 arranges a protein complex between the p97 N-domain and central channel, suggesting a hold-and-extract mechanism for p97-mediated disassembly.
- Molecular Biology I, Center of Medical Biotechnology, Faculty of Biology, University of Duisburg-Essen, Essen, Germany.
Organizational Affiliation: