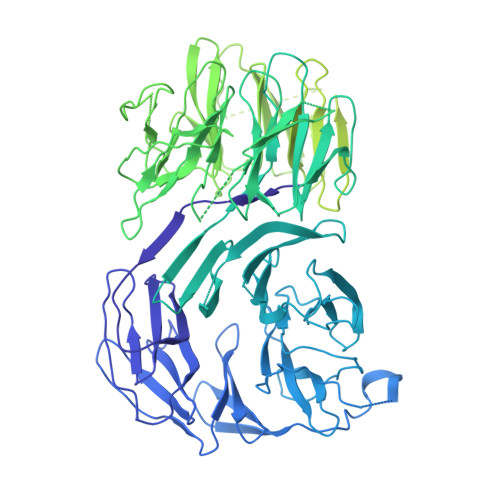
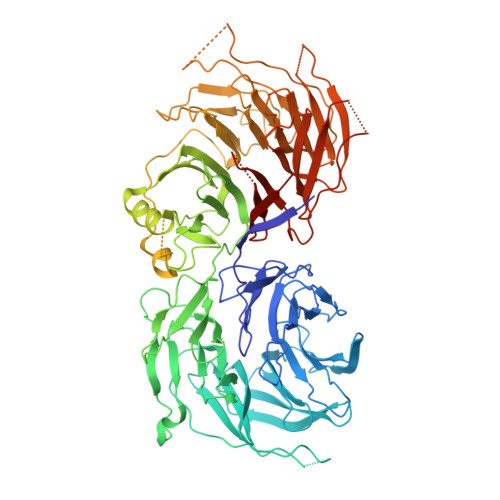
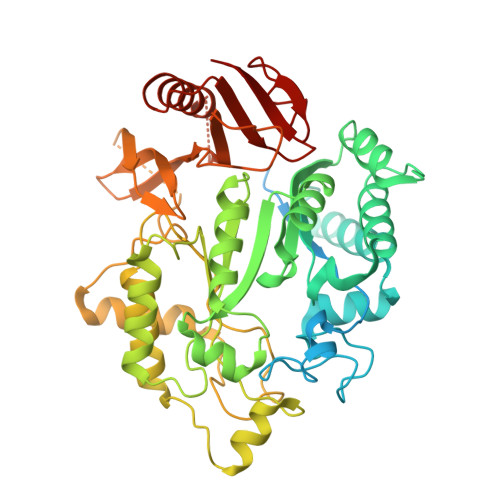
Cryo-EM structure of the fully assembled Elongator complex.
Jaciuk, M., Scherf, D., Kaszuba, K., Gaik, M., Rau, A., Koscielniak, A., Krutyholowa, R., Rawski, M., Indyka, P., Graziadei, A., Chramiec-Glabik, A., Biela, A., Dobosz, D., Lin, T.Y., Abbassi, N.E., Hammermeister, A., Rappsilber, J., Kosinski, J., Schaffrath, R., Glatt, S.(2023) Nucleic Acids Res 51: 2011-2032
- PubMed: 36617428
- DOI: https://doi.org/10.1093/nar/gkac1232
- Primary Citation of Related Structures:
8ASV, 8ASW, 8AT6, 8AVG - PubMed Abstract:
Transfer RNA (tRNA) molecules are essential to decode messenger RNA codons during protein synthesis. All known tRNAs are heavily modified at multiple positions through post-transcriptional addition of chemical groups. Modifications in the tRNA anticodons are directly influencing ribosome decoding and dynamics during translation elongation and are crucial for maintaining proteome integrity. In eukaryotes, wobble uridines are modified by Elongator, a large and highly conserved macromolecular complex. Elongator consists of two subcomplexes, namely Elp123 containing the enzymatically active Elp3 subunit and the associated Elp456 hetero-hexamer. The structure of the fully assembled complex and the function of the Elp456 subcomplex have remained elusive. Here, we show the cryo-electron microscopy structure of yeast Elongator at an overall resolution of 4.3 Å. We validate the obtained structure by complementary mutational analyses in vitro and in vivo. In addition, we determined various structures of the murine Elongator complex, including the fully assembled mouse Elongator complex at 5.9 Å resolution. Our results confirm the structural conservation of Elongator and its intermediates among eukaryotes. Furthermore, we complement our analyses with the biochemical characterization of the assembled human Elongator. Our results provide the molecular basis for the assembly of Elongator and its tRNA modification activity in eukaryotes.
- Malopolska Centre of Biotechnology (MCB), Jagiellonian University, Krakow 30-387, Poland.
Organizational Affiliation: