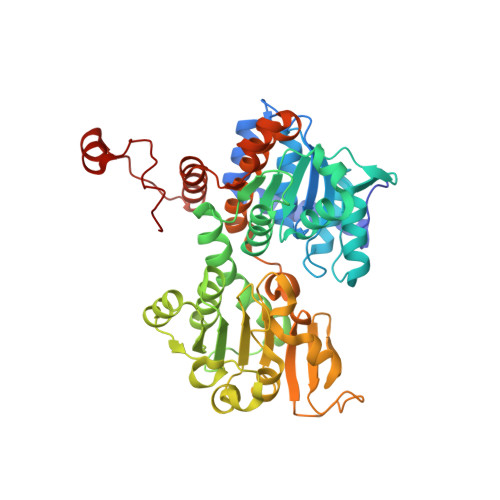
Crystal structure of the Q65A mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations
Drozdzal, P., Wozniak, K., Malecki, P., Gawel, M., Komorowska, M., Brzezinski, K.To be published.