Semisynthetic LC3 Probes for Autophagy Pathways Reveal a Noncanonical LC3 Interacting Region Motif Crucial for the Enzymatic Activity of Human ATG3.
Farnung, J., Muhar, M., Liang, J.R., Tolmachova, K.A., Benoit, R.M., Corn, J.E., Bode, J.W.(2023) ACS Cent Sci 9: 1025-1034
- PubMed: 37252361
- DOI: https://doi.org/10.1021/acscentsci.3c00009
- Primary Citation of Related Structures:
8AFI - PubMed Abstract:
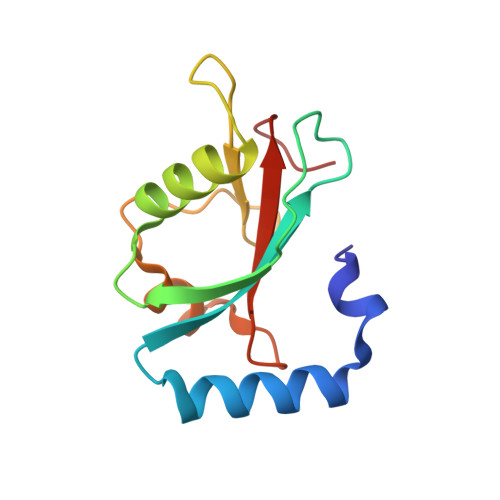
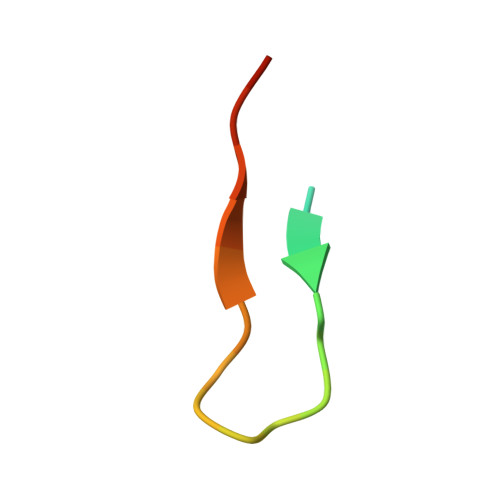
Macroautophagy is one of two major degradation systems in eukaryotic cells. Regulation and control of autophagy are often achieved through the presence of short peptide sequences called LC3 interacting regions (LIR) in autophagy-involved proteins. Using a combination of new protein-derived activity-based probes prepared from recombinant LC3 proteins, along with protein modeling and X-ray crystallography of the ATG3-LIR peptide complex, we identified a noncanonical LIR motif in the human E2 enzyme responsible for LC3 lipidation, ATG3. The LIR motif is present in the flexible region of ATG3 and adopts an uncommon β-sheet structure binding to the backside of LC3. We show that the β-sheet conformation is crucial for its interaction with LC3 and used this insight to design synthetic macrocyclic peptide-binders to ATG3. CRISPR-enabled in cellulo studies provide evidence that LIR ATG3 is required for LC3 lipidation and ATG3∼LC3 thioester formation. Removal of LIR ATG3 negatively impacts the rate of thioester transfer from ATG7 to ATG3.
- Laboratory for Organic Chemistry, Department of Chemistry and Applied Biosciences ETH Zürich, CH-8093 Zürich, Switzerland.
Organizational Affiliation: