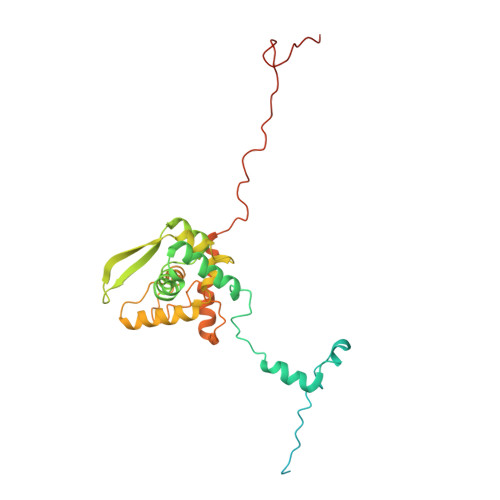
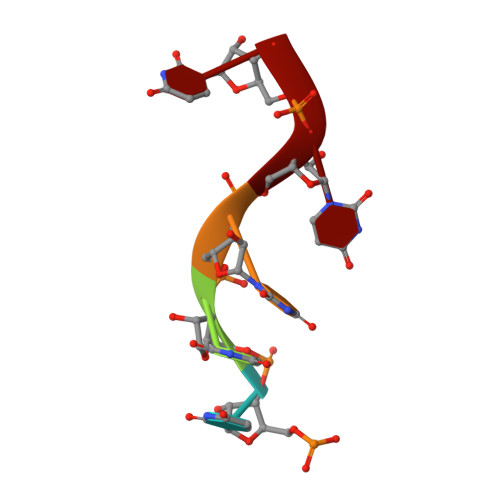
CryoEM and stability analysis of virus-like particles of potyvirus and ipomovirus infecting a common host.
Chase, O., Javed, A., Byrne, M.J., Thuenemann, E.C., Lomonossoff, G.P., Ranson, N.A., Lopez-Moya, J.J.(2023) Commun Biol 6: 433-433
- PubMed: 37076658
- DOI: https://doi.org/10.1038/s42003-023-04799-x
- Primary Citation of Related Structures:
8ACB, 8ACC - PubMed Abstract:
Sweet potato feathery mottle virus (SPFMV) and Sweet potato mild mottle virus (SPMMV) are members of the genera Potyvirus and Ipomovirus, family Potyviridae, sharing Ipomoea batatas as common host, but transmitted, respectively, by aphids and whiteflies. Virions of family members consist of flexuous rods with multiple copies of a single coat protein (CP) surrounding the RNA genome. Here we report the generation of virus-like particles (VLPs) by transient expression of the CPs of SPFMV and SPMMV in the presence of a replicating RNA in Nicotiana benthamiana. Analysis of the purified VLPs by cryo-electron microscopy, gave structures with resolutions of 2.6 and 3.0 Å, respectively, showing a similar left-handed helical arrangement of 8.8 CP subunits per turn with the C-terminus at the inner surface and a binding pocket for the encapsidated ssRNA. Despite their similar architecture, thermal stability studies reveal that SPMMV VLPs are more stable than those of SPFMV.
- Centre for Research in Agricultural Genomics (CRAG, CSIC-IRTA-UAB-UB), 08193, Cerdanyola del Vallès, Barcelona, Spain.
Organizational Affiliation: