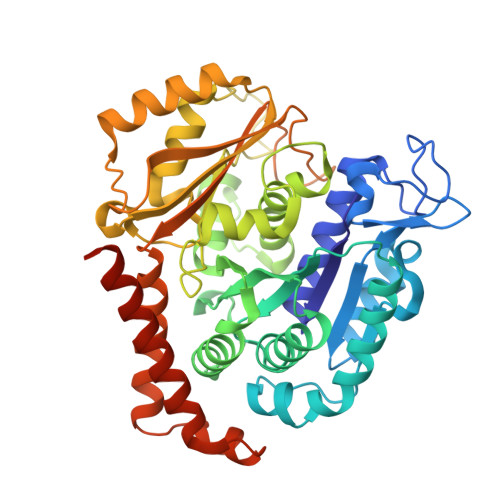
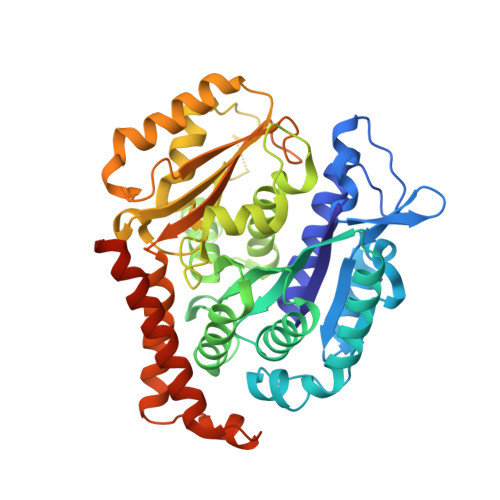
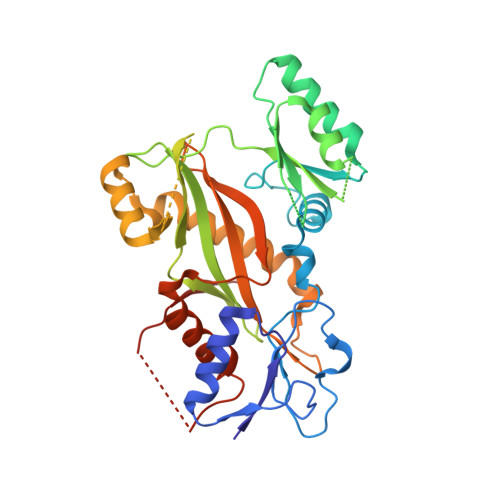
Development of [1,2]oxazoloisoindoles tubulin polymerization inhibitors: Further chemical modifications and potential therapeutic effects against lymphomas.
Barreca, M., Spano, V., Rocca, R., Bivacqua, R., Abel, A.C., Maruca, A., Montalbano, A., Raimondi, M.V., Tarantelli, C., Gaudio, E., Cascione, L., Rinaldi, A., Bai, R., Steinmetz, M.O., Prota, A.E., Alcaro, S., Hamel, E., Bertoni, F., Barraja, P.(2022) Eur J Med Chem 243: 114744-114744
- PubMed: 36242921
- DOI: https://doi.org/10.1016/j.ejmech.2022.114744
- Primary Citation of Related Structures:
8A9T, 8A9Z - PubMed Abstract:
Lymphomas are among the ten most common cancers, and, although progress has been achieved in increasing survival, there is still an unmet need for more effective therapeutic approaches, including better options for patients with refractory tumors that initially respond but then relapse. The lack of effective alternative treatment options highlights the need to develop new therapeutic strategies capable of improving survival prospects for lymphoma patients. Herein, we describe the identification and exploration of the SAR of a series of [1,2]oxazolo[5,4-e]isoindoles as potent small molecules that bind to the colchicine site of tubulin and that have promise for the treatment of refractory lymphomas. Exploration of the chemical space of this class of compounds at the pyrrole moiety and at the [1,2]oxazole ring highlighted two compounds bearing a 3,5-dimethoxybenzyl and a 3,4,5-trimethoxybenzyl group as potent candidates and showed that structural modifications at the isoxazole moiety are generally not favorable for activity. The two best candidates showed efficacy against different lymphoma histotypes and displayed 88 and 80% inhibition of colchicine binding fitting well into the colchicine pocket, as demonstrated by X-ray crystallography T 2 R-TTL-complexes, docking and thermodynamic analysis of the tubulin-colchicine complex structure. These results were confirmed by transcriptome data, thus indicating [1,2]oxazolo[5,4-e]isoindoles are promising candidates as antitubulin agents for the treatment of refractory lymphomas.
- Department of Biological, Chemical and Pharmaceutical Sciences and Technologies (STEBICEF), University of Palermo, Via Archirafi 32, 90123, Palermo, Italy.
Organizational Affiliation: