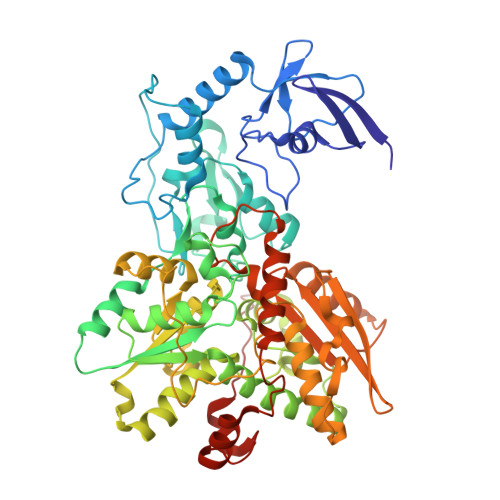
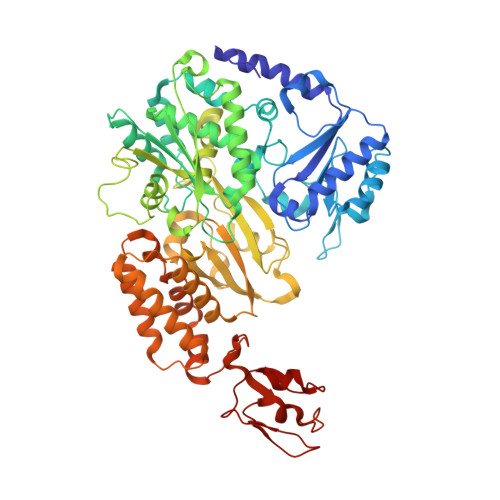
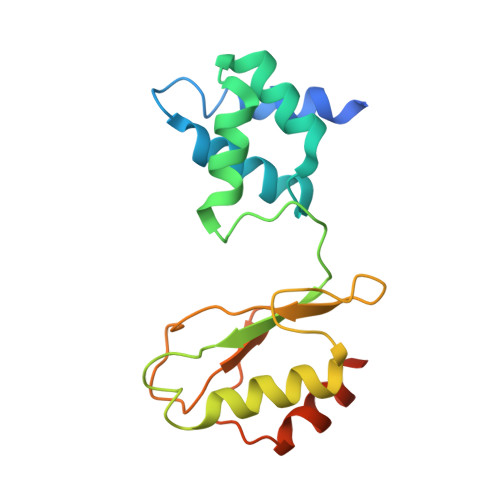
Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
Katsyv, A., Kumar, A., Saura, P., Poverlein, M.C., Freibert, S.A., T Stripp, S., Jain, S., Gamiz-Hernandez, A.P., Kaila, V.R.I., Muller, V., Schuller, J.M.(2023) J Am Chem Soc 145: 5696-5709
- PubMed: 36811855
- DOI: https://doi.org/10.1021/jacs.2c11683
- Primary Citation of Related Structures:
7Q4V, 7Q4W, 8A5E, 8A6T, 8BEW - PubMed Abstract:
Electron bifurcation is a fundamental energy coupling mechanism widespread in microorganisms that thrive under anoxic conditions. These organisms employ hydrogen to reduce CO 2 , but the molecular mechanisms have remained enigmatic. The key enzyme responsible for powering these thermodynamically challenging reactions is the electron-bifurcating [FeFe]-hydrogenase HydABC that reduces low-potential ferredoxins (Fd) by oxidizing hydrogen gas (H 2 ). By combining single-particle cryo-electron microscopy (cryoEM) under catalytic turnover conditions with site-directed mutagenesis experiments, functional studies, infrared spectroscopy, and molecular simulations, we show that HydABC from the acetogenic bacteria Acetobacterium woodii and Thermoanaerobacter kivui employ a single flavin mononucleotide (FMN) cofactor to establish electron transfer pathways to the NAD(P) + and Fd reduction sites by a mechanism that is fundamentally different from classical flavin-based electron bifurcation enzymes. By modulation of the NAD(P) + binding affinity via reduction of a nearby iron-sulfur cluster, HydABC switches between the exergonic NAD(P) + reduction and endergonic Fd reduction modes. Our combined findings suggest that the conformational dynamics establish a redox-driven kinetic gate that prevents the backflow of the electrons from the Fd reduction branch toward the FMN site, providing a basis for understanding general mechanistic principles of electron-bifurcating hydrogenases.
- Department of Molecular Microbiology & Bioenergetics, Institute of Molecular Biosciences, Johann Wolfgang Goethe University, Frankfurt am Main 60438, Germany.
Organizational Affiliation: