Crystal structure of human STING in complex with 3',3'-c-di-(2'F,2'dAMP)
Klima, M., Smola, M., Boura, E.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Stimulator of interferon protein | 204 | Homo sapiens | Mutation(s): 0 Gene Names: STING, LOC340061, hCG_1782396 | 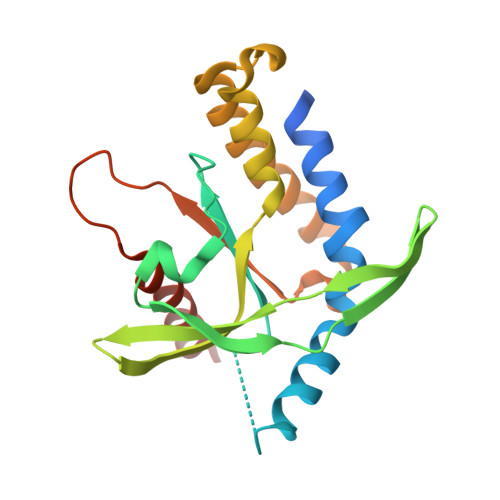 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q86WV6 (Homo sapiens) Explore Q86WV6 Go to UniProtKB: Q86WV6 | |||||
PHAROS: Q86WV6 GTEx: ENSG00000184584 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q86WV6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ubiquitin-like protein SMT3 | 98 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: SMT3, YDR510W, D9719.15 | 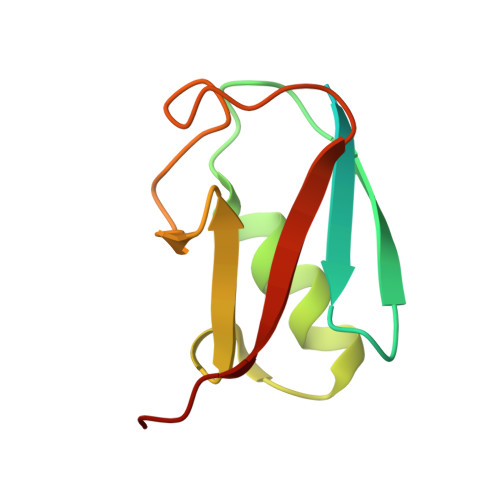 | |
UniProt | |||||
Find proteins for Q12306 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q12306 Go to UniProtKB: Q12306 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q12306 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| K43 (Subject of Investigation/LOI) Query on K43 | D [auth A] | 9-[(1~{R},6~{R},8~{R},9~{S},10~{R},15~{R},17~{R},18~{S})-17-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,11,13-tetraoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-8-yl]purin-6-amine C22 H26 F2 N10 O8 P2 WIKMGYCWQYAWTF-SSMVINTDSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 92.969 | α = 90 |
| b = 92.969 | β = 90 |
| c = 203.905 | γ = 120 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |
| Coot | model building |
| PHENIX | refinement |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |