KtrAB complex
Stautz, J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ktr system potassium uptake protein A | 220 | Vibrio alginolyticus | Mutation(s): 0 Gene Names: ktrA Membrane Entity: Yes | 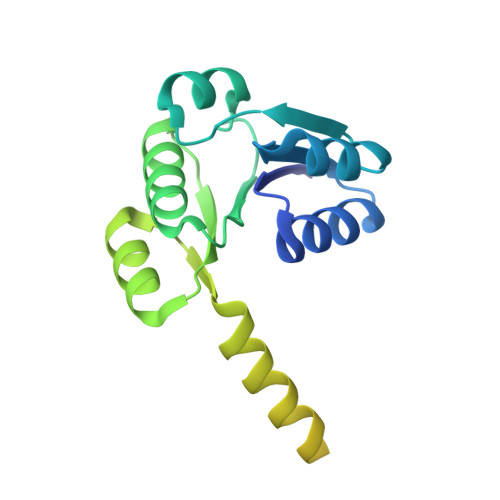 | |
UniProt | |||||
Find proteins for O87952 (Vibrio alginolyticus) Explore O87952 Go to UniProtKB: O87952 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O87952 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ktr system potassium uptake protein B | E [auth I], J [auth M], K [auth C], L [auth G] | 455 | Vibrio alginolyticus | Mutation(s): 0 Gene Names: ktrB Membrane Entity: Yes | 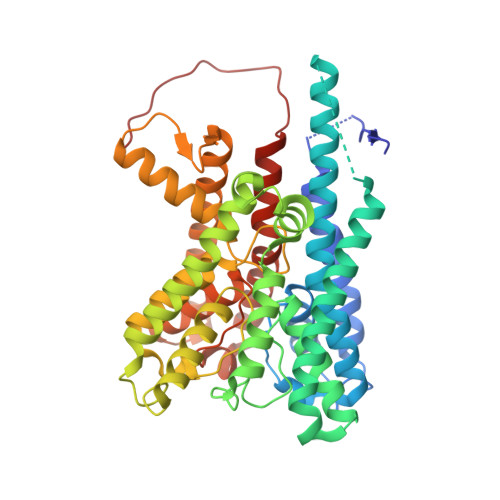 |
UniProt | |||||
Find proteins for O87953 (Vibrio alginolyticus) Explore O87953 Go to UniProtKB: O87953 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O87953 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| LMT Query on LMT | AA [auth I] AB [auth C] BA [auth I] BB [auth C] CA [auth I] | DODECYL-BETA-D-MALTOSIDE C24 H46 O11 NLEBIOOXCVAHBD-QKMCSOCLSA-N |  | ||
| ADP Query on ADP | GA [auth A] IA [auth B] KA [auth J] MA [auth L] N [auth H] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| K Query on K | HB [auth G], OA [auth M], V [auth I], XA [auth C] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| MG Query on MG | FA [auth A] JA [auth B] LA [auth J] M [auth H] NA [auth L] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Research Foundation (DFG) | Germany | VO 1449_1-1 |
| German Research Foundation (DFG) | Germany | HA 6322/4-1 |