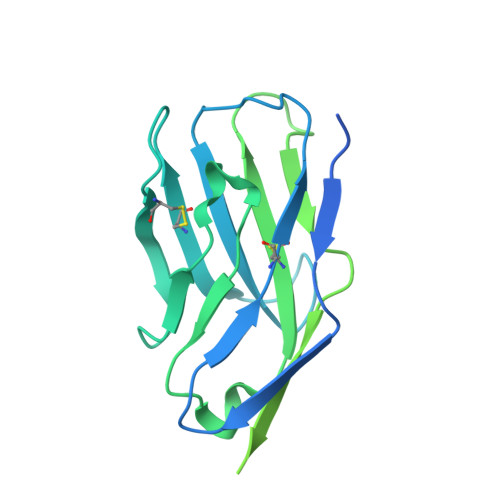
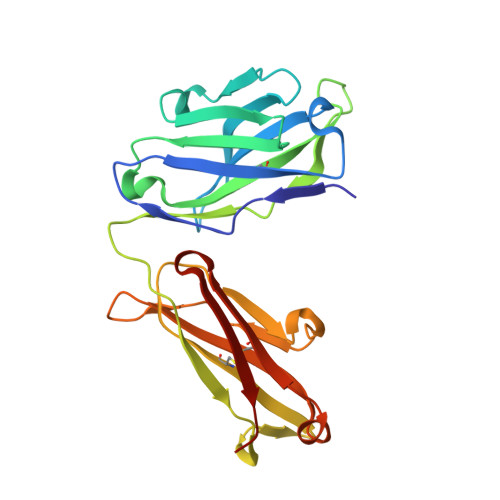
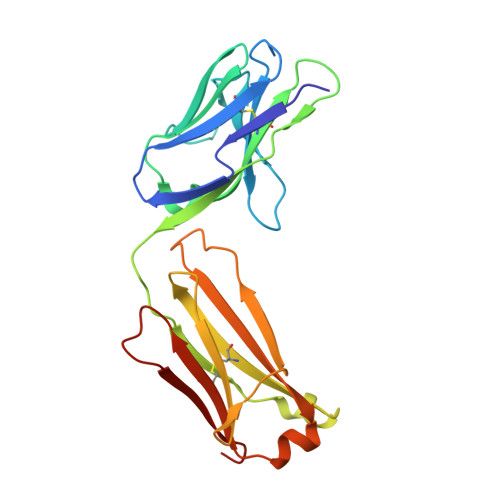
Structural insights into Siglec-15 reveal glycosylation dependency for its interaction with T cells through integrin CD11b.
Lenza, M.P., Egia-Mendikute, L., Antonana-Vildosola, A., Soares, C.O., Coelho, H., Corzana, F., Bosch, A., Manisha, P., Quintana, J.I., Oyenarte, I., Unione, L., Moure, M.J., Azkargorta, M., Atxabal, U., Sobczak, K., Elortza, F., Sutherland, J.D., Barrio, R., Marcelo, F., Jimenez-Barbero, J., Palazon, A., Ereno-Orbea, J.(2023) Nat Commun 14: 3496-3496
- PubMed: 37311743
- DOI: https://doi.org/10.1038/s41467-023-39119-8
- Primary Citation of Related Structures:
7ZOR, 7ZOZ - PubMed Abstract:
Sialic acid-binding Ig-like lectin 15 (Siglec-15) is an immune modulator and emerging cancer immunotherapy target. However, limited understanding of its structure and mechanism of action restrains the development of drug candidates that unleash its full therapeutic potential. In this study, we elucidate the crystal structure of Siglec-15 and its binding epitope via co-crystallization with an anti-Siglec-15 blocking antibody. Using saturation transfer-difference nuclear magnetic resonance (STD-NMR) spectroscopy and molecular dynamics simulations, we reveal Siglec-15 binding mode to α(2,3)- and α(2,6)-linked sialic acids and the cancer-associated sialyl-Tn (STn) glycoform. We demonstrate that binding of Siglec-15 to T cells, which lack STn expression, depends on the presence of α(2,3)- and α(2,6)-linked sialoglycans. Furthermore, we identify the leukocyte integrin CD11b as a Siglec-15 binding partner on human T cells. Collectively, our findings provide an integrated understanding of the structural features of Siglec-15 and emphasize glycosylation as a crucial factor in controlling T cell responses.
- Chemical Glycobiology lab, Center for Cooperative Research in Biosciences (CIC bioGUNE), Basque Research and Technology Alliance (BRTA), Bizkaia Technology Park, Building 800, 48160, Derio, Bizkaia, Spain.
Organizational Affiliation: