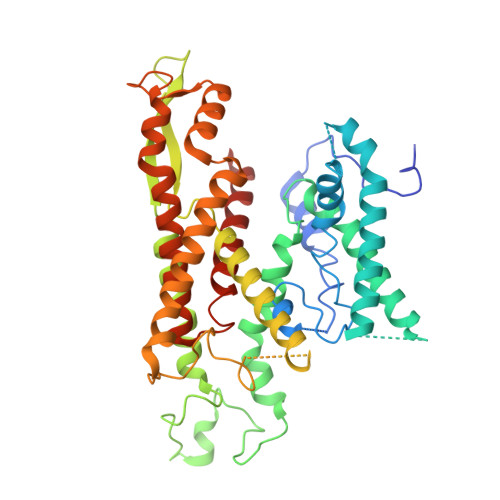
Signal peptide mimicry primes Sec61 for client-selective inhibition.
Rehan, S., Tranter, D., Sharp, P.P., Craven, G.B., Lowe, E., Anderl, J.L., Muchamuel, T., Abrishami, V., Kuivanen, S., Wenzell, N.A., Jennings, A., Kalyanaraman, C., Strandin, T., Javanainen, M., Vapalahti, O., Jacobson, M.P., McMinn, D., Kirk, C.J., Huiskonen, J.T., Taunton, J., Paavilainen, V.O.(2023) Nat Chem Biol 19: 1054-1062
- PubMed: 37169961
- DOI: https://doi.org/10.1038/s41589-023-01326-1
- Primary Citation of Related Structures:
7ZL3 - PubMed Abstract:
Preventing the biogenesis of disease-relevant proteins is an attractive therapeutic strategy, but attempts to target essential protein biogenesis factors have been hampered by excessive toxicity. Here we describe KZR-8445, a cyclic depsipeptide that targets the Sec61 translocon and selectively disrupts secretory and membrane protein biogenesis in a signal peptide-dependent manner. KZR-8445 potently inhibits the secretion of pro-inflammatory cytokines in primary immune cells and is highly efficacious in a mouse model of rheumatoid arthritis. A cryogenic electron microscopy structure reveals that KZR-8445 occupies the fully opened Se61 lateral gate and blocks access to the lumenal plug domain. KZR-8445 binding stabilizes the lateral gate helices in a manner that traps select signal peptides in the Sec61 channel and prevents their movement into the lipid bilayer. Our results establish a framework for the structure-guided discovery of novel therapeutics that selectively modulate Sec61-mediated protein biogenesis.
- Institute of Biotechnology, HiLIFE, University of Helsinki, Helsinki, Finland.
Organizational Affiliation: