Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization.
Morichaud, Z., Trapani, S., Vishwakarma, R.K., Chaloin, L., Lionne, C., Lai-Kee-Him, J., Bron, P., Brodolin, K.(2023) Nat Commun 14: 484-484
- PubMed: 36717560
- DOI: https://doi.org/10.1038/s41467-023-36113-y
- Primary Citation of Related Structures:
7ZF2 - PubMed Abstract:
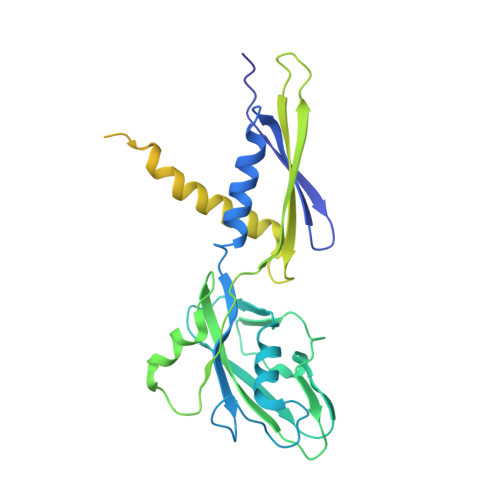
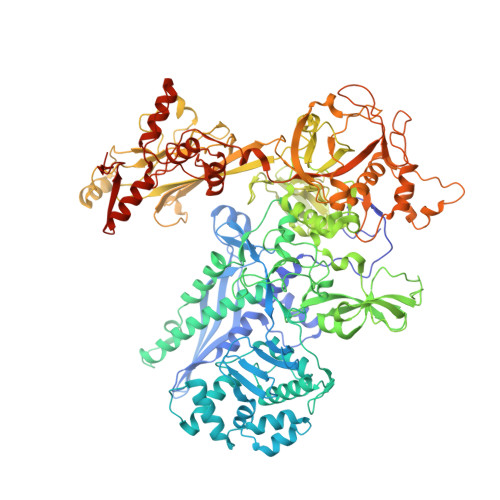
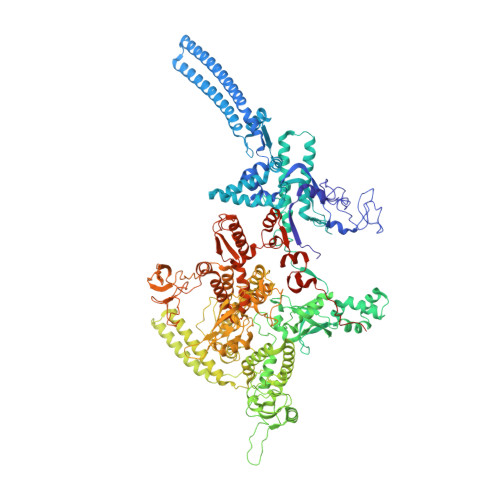
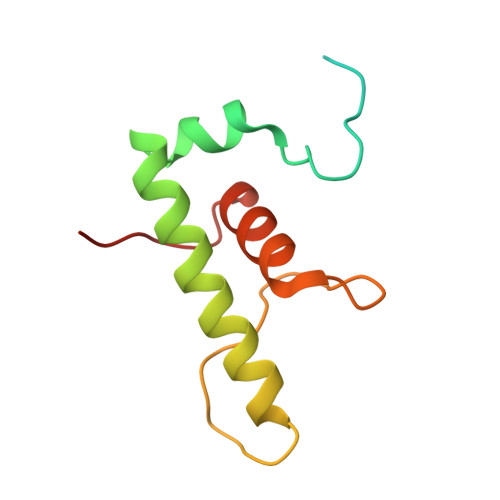
Self-assembly of macromolecules into higher-order symmetric structures is fundamental for the regulation of biological processes. Higher-order symmetric structure self-assembly by the gene expression machinery, such as bacterial DNA-dependent RNA polymerase (RNAP), has never been reported before. Here, we show that the stress-response σ B factor from the human pathogen, Mycobacterium tuberculosis, induces the RNAP holoenzyme oligomerization into a supramolecular complex composed of eight RNAP units. Cryo-electron microscopy revealed a pseudo-symmetric structure of the RNAP octamer in which RNAP protomers are captured in an auto-inhibited state and display an open-clamp conformation. The structure shows that σ B is sequestered by the RNAP flap and clamp domains. The transcriptional activator RbpA prevented octamer formation by promoting the initiation-competent RNAP conformation. Our results reveal that a non-conserved region of σ is an allosteric controller of transcription initiation and demonstrate how basal transcription factors can regulate gene expression by modulating the RNAP holoenzyme assembly and hibernation.
- Institut de Recherche en Infectiologie de Montpellier, Univ Montpellier, CNRS, Montpellier, 34293, France.
Organizational Affiliation: