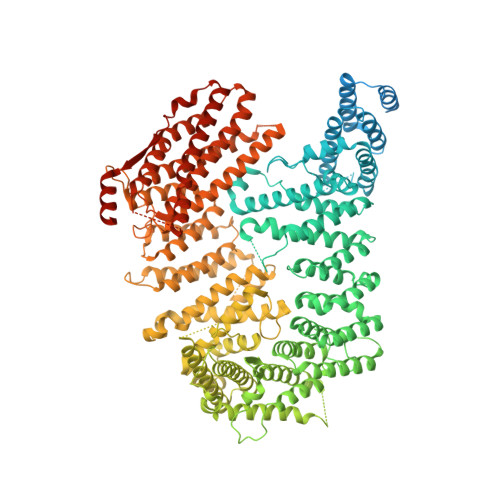
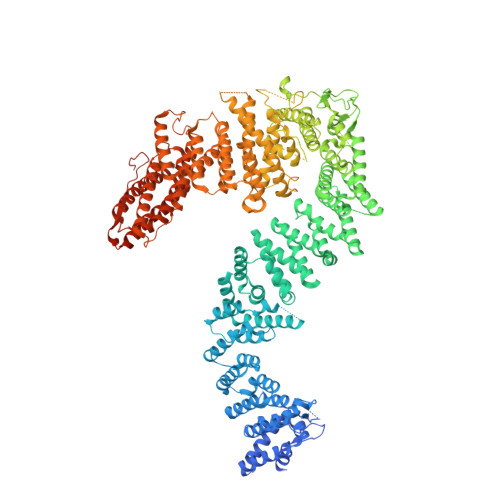
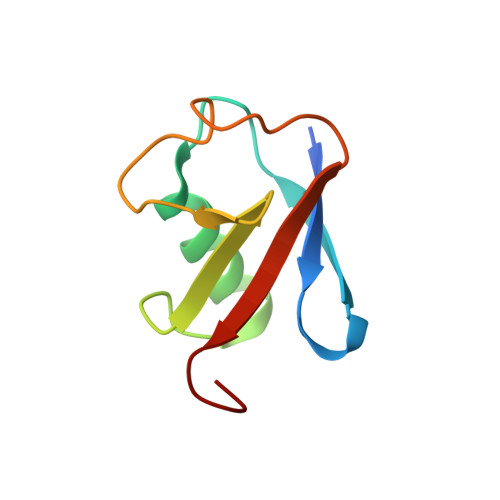
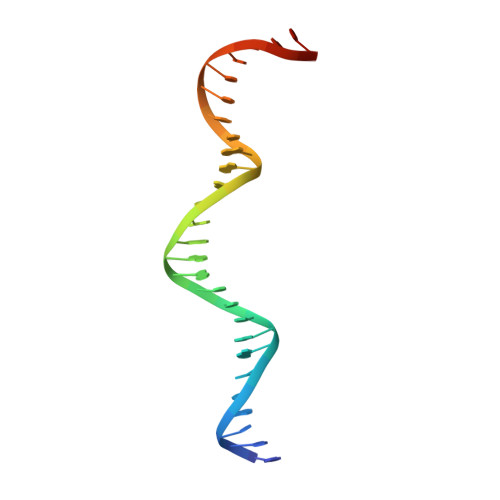
Structural and biochemical basis of interdependent FANCI-FANCD2 ubiquitination.
Lemonidis, K., Rennie, M.L., Arkinson, C., Chaugule, V.K., Clarke, M., Streetley, J., Walden, H.(2023) EMBO J 42: e111898-e111898
- PubMed: 36385258
- DOI: https://doi.org/10.15252/embj.2022111898
- Primary Citation of Related Structures:
7ZF1 - PubMed Abstract:
Di-monoubiquitination of the FANCI-FANCD2 (ID2) complex is a central and crucial step for the repair of DNA interstrand crosslinks via the Fanconi anaemia pathway. While FANCD2 ubiquitination precedes FANCI ubiquitination, FANCD2 is also deubiquitinated at a faster rate than FANCI, which can result in a FANCI-ubiquitinated ID2 complex (I Ub D2). Here, we present a 4.1 Å cryo-EM structure of I Ub D2 complex bound to double-stranded DNA. We show that this complex, like ID2 Ub and I Ub D2 Ub , is also in the closed ID2 conformation and clamps on DNA. The target lysine of FANCD2 (K561) becomes fully exposed in the I Ub D2-DNA structure and is thus primed for ubiquitination. Similarly, FANCI's target lysine (K523) is also primed for ubiquitination in the ID2 Ub -DNA complex. The I Ub D2-DNA complex exhibits deubiquitination resistance, conferred by the presence of DNA and FANCD2. ID2 Ub -DNA, on the other hand, can be efficiently deubiquitinated by USP1-UAF1, unless further ubiquitination on FANCI occurs. Therefore, FANCI ubiquitination effectively maintains FANCD2 ubiquitination in two ways: it prevents excessive FANCD2 deubiquitination within an I Ub D2 Ub -DNA complex, and it enables re-ubiquitination of FANCD2 within a transient, closed-on-DNA, I Ub D2 complex.
- School of Molecular Biosciences, College of Medical Veterinary and Life Sciences, University of Glasgow, Glasgow, UK.
Organizational Affiliation: