Full-length structure of a FIC effector protein from Bartonella reveals a novel structural domain
Huber, M., Wagner, A., Reiners, J., Seyfert, C.E.M., Smits, S.H.J., Schirmer, T., Dehio, C.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Bartonella effector protein (Bep) substrate of VirB T4SS | 566 | Bartonella clarridgeiae | Mutation(s): 0 Gene Names: BARCL_0069 EC: 2.7.7.108 | 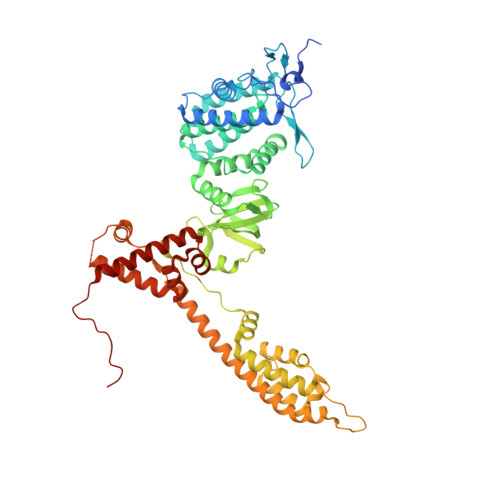 | |
UniProt | |||||
Find proteins for E6YFW2 (Bartonella clarridgeiae (strain CCUG 45776 / CIP 104772 / 73)) Explore E6YFW2 Go to UniProtKB: E6YFW2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | E6YFW2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 70.492 | α = 90 |
| b = 70.492 | β = 90 |
| c = 213.961 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Aimless | data scaling |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Swiss National Science Foundation | Switzerland | -- |