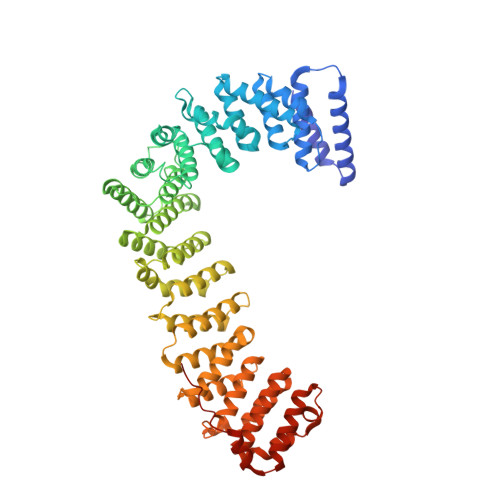
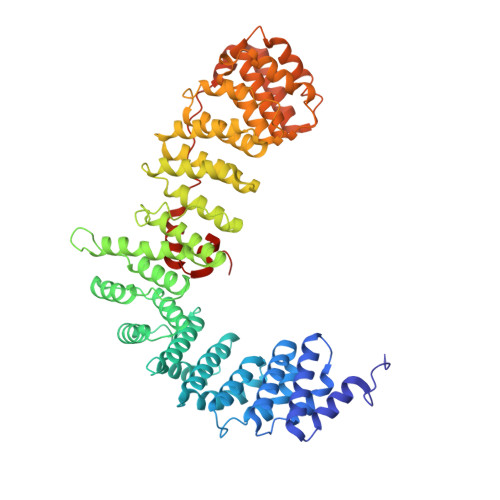
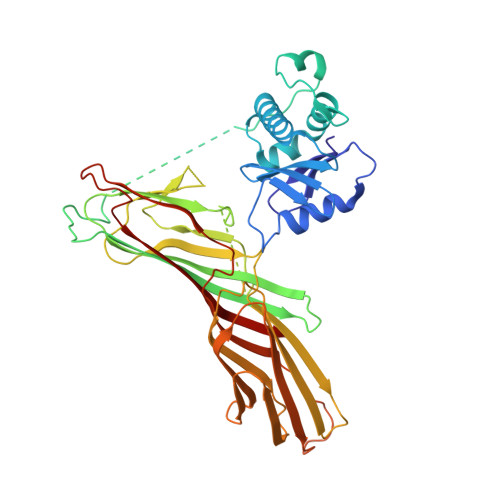
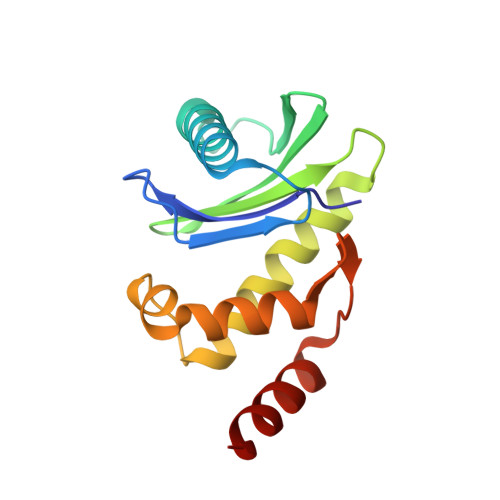
FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P 2 -dependent switch.
Zaccai, N.R., Kadlecova, Z., Dickson, V.K., Korobchevskaya, K., Kamenicky, J., Kovtun, O., Umasankar, P.K., Wrobel, A.G., Kaufman, J.G.G., Gray, S.R., Qu, K., Evans, P.R., Fritzsche, M., Sroubek, F., Honing, S., Briggs, J.A.G., Kelly, B.T., Owen, D.J., Traub, L.M.(2022) Sci Adv 8: eabn2018-eabn2018
- PubMed: 35486718
- DOI: https://doi.org/10.1126/sciadv.abn2018
- Primary Citation of Related Structures:
7OFP, 7OG1, 7OHI, 7OHO, 7OHZ, 7OI5, 7OIQ, 7OIT, 7Z5C - PubMed Abstract:
Clathrin-mediated endocytosis (CME) is the main mechanism by which mammalian cells control their cell surface proteome. Proper operation of the pivotal CME cargo adaptor AP2 requires membrane-localized Fer/Cip4 homology domain-only proteins (FCHO). Here, live-cell enhanced total internal reflection fluorescence-structured illumination microscopy shows that FCHO marks sites of clathrin-coated pit (CCP) initiation, which mature into uniform-sized CCPs comprising a central patch of AP2 and clathrin corralled by an FCHO/Epidermal growth factor potential receptor substrate number 15 (Eps15) ring. We dissect the network of interactions between the FCHO interdomain linker and AP2, which concentrates, orients, tethers, and partially destabilizes closed AP2 at the plasma membrane. AP2's subsequent membrane deposition drives its opening, which triggers FCHO displacement through steric competition with phosphatidylinositol 4,5-bisphosphate, clathrin, cargo, and CME accessory factors. FCHO can now relocate toward a CCP's outer edge to engage and activate further AP2s to drive CCP growth/maturation.
- CIMR, University of Cambridge, Biomedical Campus, Hills Road, Cambridge CB2 0XY, UK.
Organizational Affiliation: