Novel fragment-derived colchicine-site binders as microtubule-destabilizing agents.
de la Roche, N.M., Muhlethaler, T., Di Martino, R.M.C., Ortega, J.A., Gioia, D., Roy, B., Prota, A.E., Steinmetz, M.O., Cavalli, A.(2022) Eur J Med Chem 241: 114614-114614
- PubMed: 35939994
- DOI: https://doi.org/10.1016/j.ejmech.2022.114614
- Primary Citation of Related Structures:
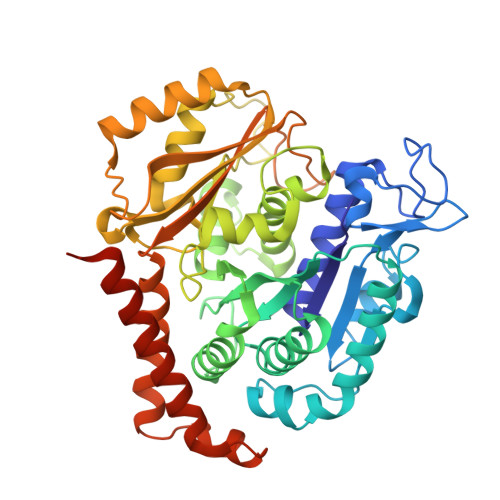
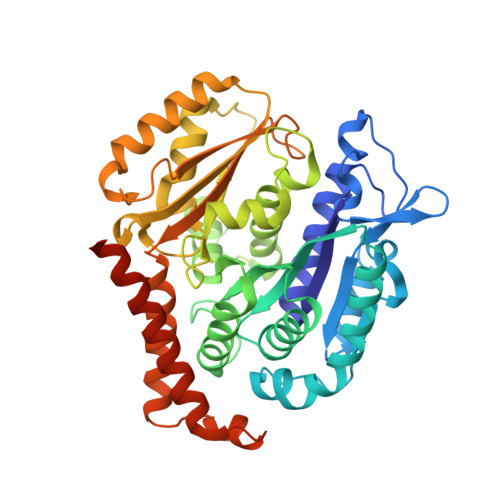
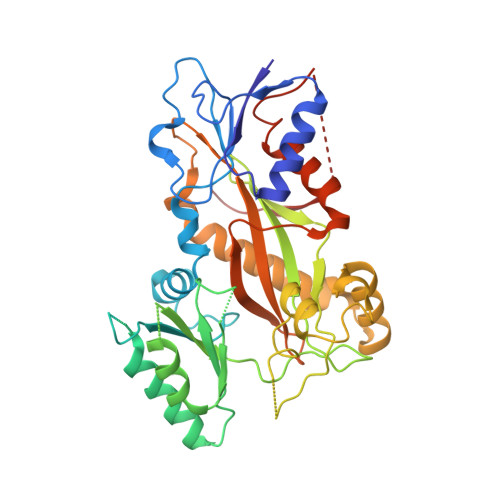
7Z2N, 7Z2P - PubMed Abstract:
Microtubules (MTs) are dynamic filaments of the cytoskeleton, which are formed by the polymerization of their building block tubulin. Perturbation of MT dynamics by MT-targeting agents (MTAs) leads to cell cycle arrest or cell death, a strategy that is pursued in chemotherapy. We recently performed a combined computational and crystallographic fragment screening approach and identified several tubulin-binding fragments. Here, we sought to capitalize on this study with the aim to demonstrate that low affinity tubulin-binding fragments can indeed be used as valuable starting points for the development of active, lead-like antitubulin small molecules. To this end, we report on a new, rationally designed series of 2-aminobenzimidazole derivatives that destabilize MTs by binding tubulin at the colchicine-binding site (CBS). We applied a fragment growing strategy by combining X-ray crystallography and computer-aided drug design. Preliminary structure-activity-relationship studies afforded compound 18 that inhibits HeLa cell viability with a submicromolar activity (IC 50 of 0.9 μM). X-ray crystallography confirmed the compound pose in the CBS, while immunostaining experiments suggested a molecular mechanism of action alike classical CBS ligands with antimitotic and antitumor activity associated with MTs destabilization. This promising outcome underpins that our previously performed combined computational and crystallographic fragment screening approach provides promising starting points for developing new MTAs binding to the CBS of tubulin and, eventually, to further tubulin pockets.
- Computational & Chemical Biology, Istituto Italiano di Tecnologia, Via Morego, 30, 16163, Genova, Italy; Department of Pharmacy and Biotechnology, Alma Mater Studiorum, University of Bologna, Via Belmeloro 6, 40126, Bologna, Italy.
Organizational Affiliation: