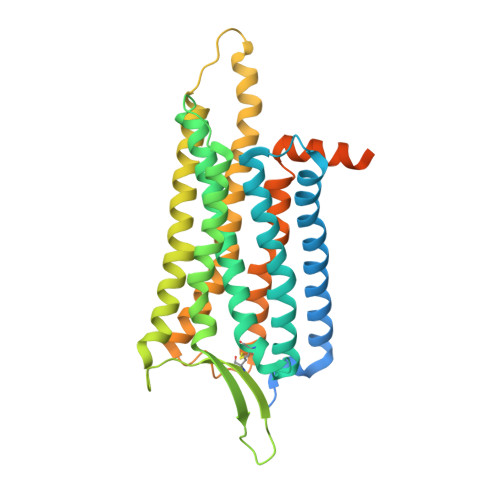
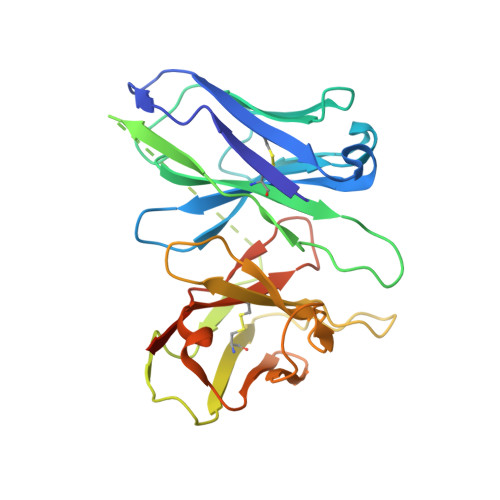
Structural basis for Y2 receptor-mediated neuropeptide Y and peptide YY signaling.
Kang, H., Park, C., Choi, Y.K., Bae, J., Kwon, S., Kim, J., Choi, C., Seok, C., Im, W., Choi, H.J.(2023) Structure 31: 44-57.e6
- PubMed: 36525977
- DOI: https://doi.org/10.1016/j.str.2022.11.010
- Primary Citation of Related Structures:
7YON, 7YOO - PubMed Abstract:
Neuropeptide Y (NPY) and its receptors are expressed in various human tissues including the brain where they regulate appetite and emotion. Upon NPY stimulation, the neuropeptide Y1 and Y2 receptors (Y 1 R and Y 2 R, respectively) activate G I signaling, but their physiological responses to food intake are different. In addition, deletion of the two N-terminal amino acids of peptide YY (PYY(3-36)), the endogenous form found in circulation, can stimulate Y 2 R but not Y 1 R, suggesting that Y 1 R and Y 2 R may have distinct ligand-binding modes. Here, we report the cryo-electron microscopy structures of the PYY(3-36)‒Y 2 R‒G i and NPY‒Y 2 R‒G i complexes. Using cell-based assays, molecular dynamics simulations, and structural analysis, we revealed the molecular basis of the exclusive binding of PYY(3-36) to Y 2 R. Furthermore, we demonstrated that Y 2 R favors G protein signaling over β-arrestin signaling upon activation, whereas Y 1 R does not show a preference between these two pathways.
- Department of Biological Sciences, Seoul National University, Seoul 08826, Republic of Korea.
Organizational Affiliation: