Cryo-EM structure of the human chemerin receptor 1-Gi protein complex bound to the C-terminal nonapeptide of chemerin.
Wang, J., Chen, G., Liao, Q., Lyu, W., Liu, A., Zhu, L., Du, Y., Ye, R.D.(2023) Proc Natl Acad Sci U S A 120: e2214324120-e2214324120
- PubMed: 36881626
- DOI: https://doi.org/10.1073/pnas.2214324120
- Primary Citation of Related Structures:
7YKD - PubMed Abstract:
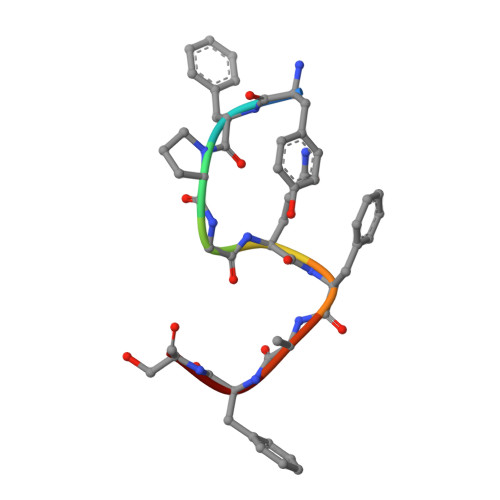
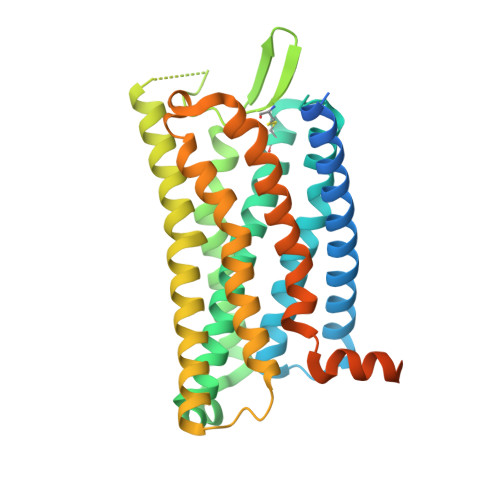
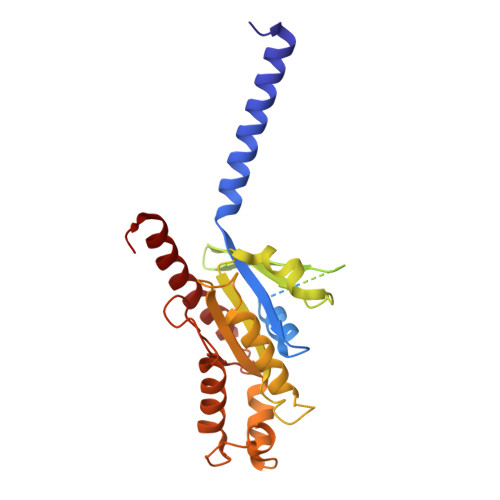
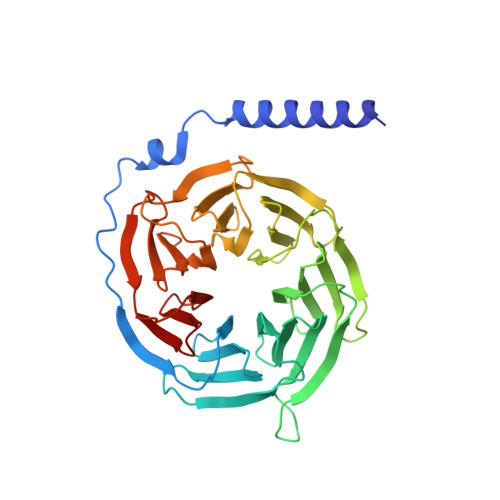
Chemerin is a processed protein that acts on G protein-coupled receptors (GPCRs) for its chemotactic and adipokine activities. The biologically active chemerin (chemerin 21-157) results from proteolytic cleavage of prochemerin and uses its C-terminal peptide containing the sequence YFPGQFAFS for receptor activation. Here we report a high-resolution cryo-electron microscopy (cryo-EM) structure of human chemerin receptor 1 (CMKLR1) bound to the C-terminal nonapeptide of chemokine (C9) in complex with Gi proteins. C9 inserts its C terminus into the binding pocket and is stabilized through hydrophobic interactions involving its Y1, F2, F6, and F8, as well as polar interactions between G4, S9, and several amino acids lining the binding pocket of CMKLR1. Microsecond scale molecular dynamics simulations support a balanced force distribution across the whole ligand-receptor interface that enhances thermodynamic stability of the captured binding pose of C9. The C9 interaction with CMKLR1 is drastically different from chemokine recognition by chemokine receptors, which follow a two-site two-step model. In contrast, C9 takes an "S"-shaped pose in the binding pocket of CMKLR1 much like angiotensin II in the AT1 receptor. Our mutagenesis and functional analyses confirmed the cryo-EM structure and key residues in the binding pocket for these interactions. Our findings provide a structural basis for chemerin recognition by CMKLR1 for the established chemotactic and adipokine activities.
- Kobilka Institute of Innovative Drug Discovery, School of Medicine, The Chinese University of Hong Kong, Shenzhen, Guangdong 518172, P.R. China.
Organizational Affiliation: