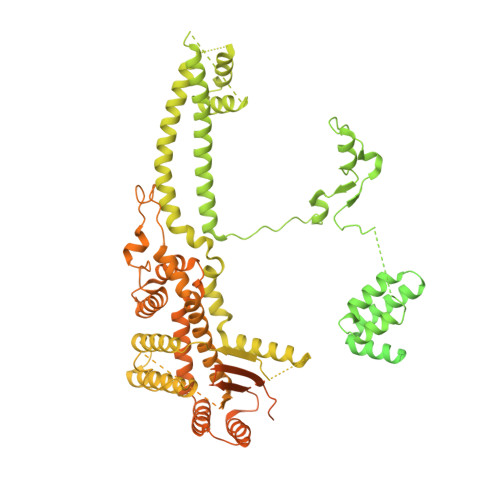
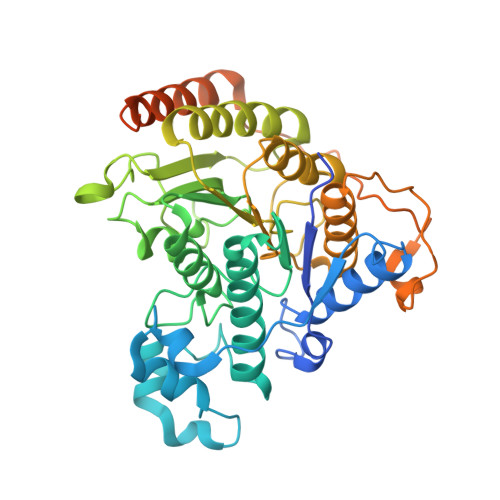
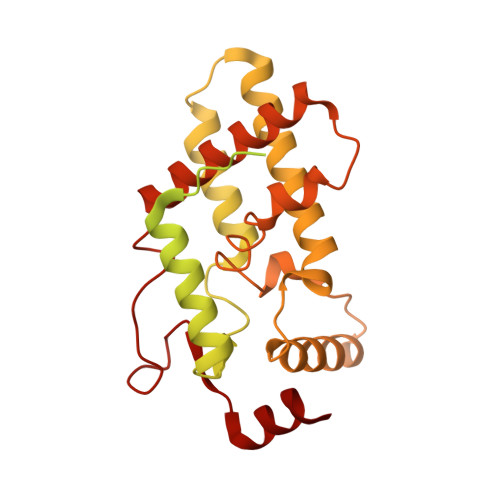
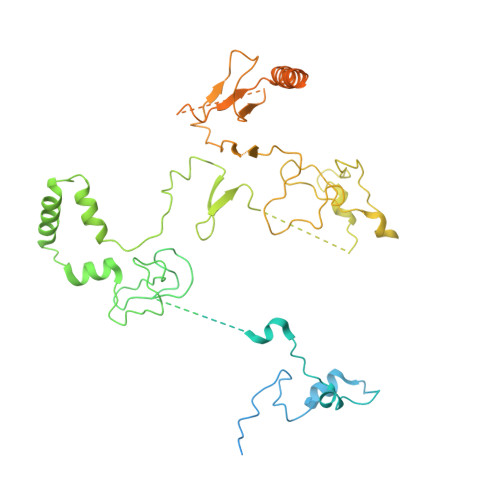
Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Guan, H., Wang, P., Zhang, P., Ruan, C., Ou, Y., Peng, B., Zheng, X., Lei, J., Li, B., Yan, C., Li, H.(2023) Nature 620: 669-675
- PubMed: 37468628
- DOI: https://doi.org/10.1038/s41586-023-06349-1
- Primary Citation of Related Structures:
7YI0, 7YI1, 7YI2, 7YI3, 7YI4, 7YI5 - PubMed Abstract:
Context-dependent dynamic histone modifications constitute a key epigenetic mechanism in gene regulation 1-4 . The Rpd3 small (Rpd3S) complex recognizes histone H3 trimethylation on lysine 36 (H3K36me3) and deacetylates histones H3 and H4 at multiple sites across transcribed regions 5-7 . Here we solved the cryo-electron microscopy structures of Saccharomyces cerevisiae Rpd3S in its free and H3K36me3 nucleosome-bound states. We demonstrated a unique architecture of Rpd3S, in which two copies of Eaf3-Rco1 heterodimers are asymmetrically assembled with Rpd3 and Sin3 to form a catalytic core complex. Multivalent recognition of two H3K36me3 marks, nucleosomal DNA and linker DNAs by Eaf3, Sin3 and Rco1 positions the catalytic centre of Rpd3 next to the histone H4 N-terminal tail for deacetylation. In an alternative catalytic mode, combinatorial readout of unmethylated histone H3 lysine 4 and H3K36me3 by Rco1 and Eaf3 directs histone H3-specific deacetylation except for the registered histone H3 acetylated lysine 9. Collectively, our work illustrates dynamic and diverse modes of multivalent nucleosomal engagement and methylation-guided deacetylation by Rpd3S, highlighting the exquisite complexity of epigenetic regulation with delicately designed multi-subunit enzymatic machineries in transcription and beyond.
- State Key Laboratory of Molecular Oncology, MOE Key Laboratory of Protein Sciences, SXMU-Tsinghua Collaborative Innovation Center for Frontier Medicine, School of Medicine, Tsinghua University, Beijing, China.
Organizational Affiliation: