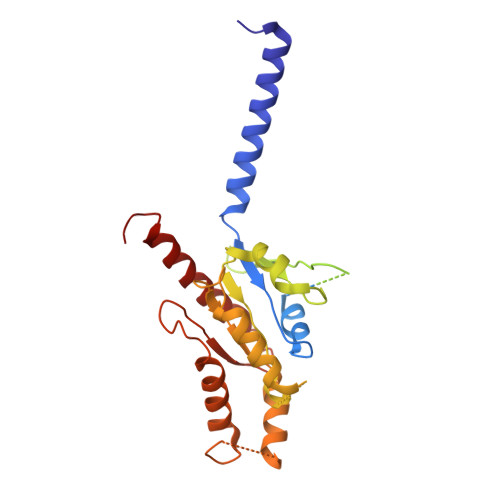
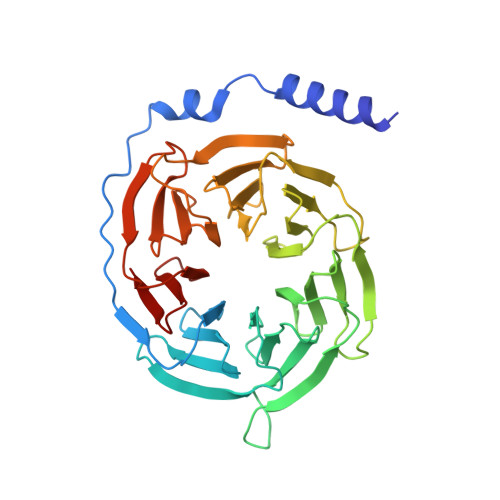
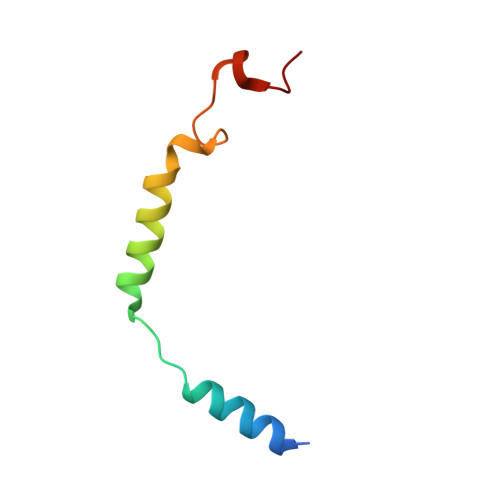
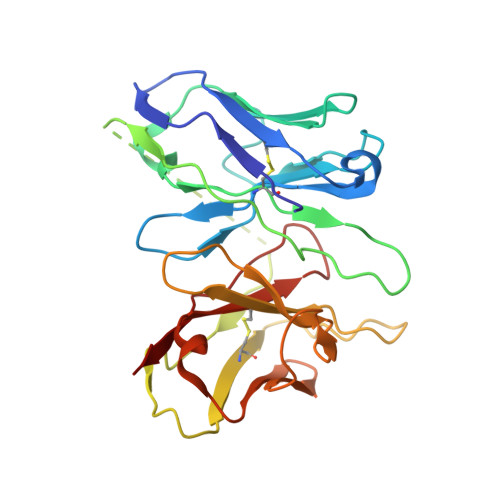
Cryo-EM structure of G-protein-coupled receptor GPR17 in complex with inhibitory G protein.
Ye, F., Wong, T.S., Chen, G., Zhang, Z., Zhang, B., Gan, S., Gao, W., Li, J., Wu, Z., Pan, X., Du, Y.(2022) MedComm (2020) 3: e159-e159
- PubMed: 36105372
- DOI: https://doi.org/10.1002/mco2.159
- Primary Citation of Related Structures:
7Y89 - PubMed Abstract:
GPR17 is a class A orphan G protein-coupled receptor (GPCR) expressed in neurons and oligodendrocyte progenitors of the central nervous system (CNS). The signalling of GPR17 occurs through the heterotrimeric Gi, but its activation mechanism is unclear. Here, we employed cryo-electron microscopy (cryo-EM) technology to elucidate the structure of activated GPR17-Gi complex. The 3.02 Å resolution structure, together with mutagenesis studies, revealed that the extracellular loop2 of GPR17 occupied the orthosteric binding pocket to promote its self-activation. The active GPR17 carried several typical microswitches like other class A GPCRs. Moreover, the Gi interacted with the key residues of transmembrane helix 3 (TM3), the amphipathic helix 8 (Helix8), and intracellular loops 3 (ICL3) in GPR17 to engage in the receptor core. In summary, our results highlight the activation mechanism of GPR17 from the structural basis. Elucidating the structural and activation mechanism of GPR17 may facilitate the pharmacological intervention for acute/chronic CNS injury.
- Kobilka Institute of Innovative Drug Discovery Shenzhen Key Laboratory of Steroid Drug Discovery and Development School of Medicine The Chinese University of Hong Kong Shenzhen Guangdong China.
Organizational Affiliation: