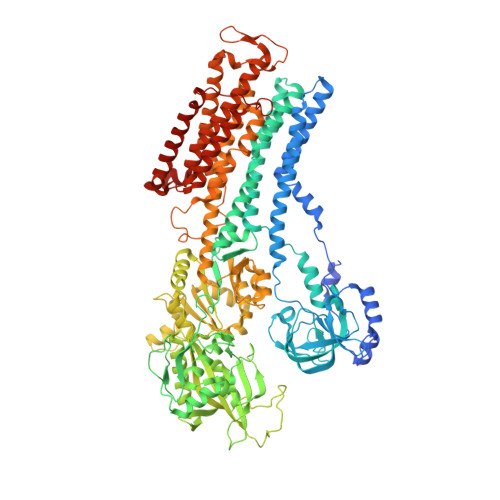
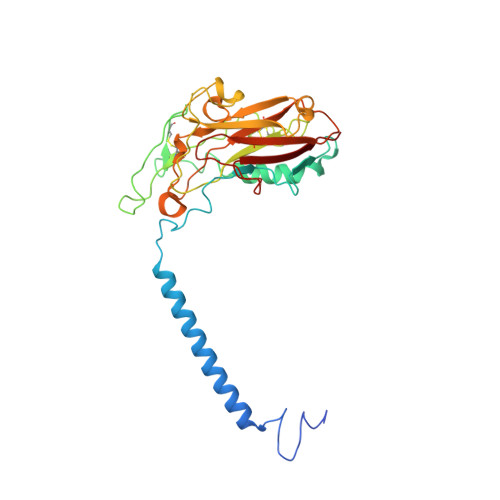
Cryo-electron microscopy of Na + ,K + -ATPase reveals how the extracellular gate locks in the E2·2K + state.
Kanai, R., Cornelius, F., Vilsen, B., Toyoshima, C.(2022) FEBS Lett 596: 2513-2524
- PubMed: 35747985
- DOI: https://doi.org/10.1002/1873-3468.14437
- Primary Citation of Related Structures:
7Y45, 7Y46 - PubMed Abstract:
Na + ,K + -ATPase (NKA) is one of the most important members of the P-type ion-translocating ATPases and plays a pivotal role in establishing electrochemical gradients for Na + and K + across the cell membrane. Presented here is a 3.3 Å resolution structure of NKA in the E2·2K + state solved by cryo-electron microscopy. It is a stable state with two occluded K + after transferring three Na + into the extracellular medium and releasing inorganic phosphate bound to the cytoplasmic P domain. We describe how the extracellular ion pathway wide open in the E2P state becomes closed and locked in E2·2K + , linked to events at the phosphorylation site more than 50 Å away. We also show, although at low resolution, how ATP binding to NKA in E2·2K + relaxes the gating machinery and thereby accelerates the transition into the next step, that is, the release of K + into the cytoplasm, more than 100 times.
- Institute for Quantitative Biosciences, The University of Tokyo, Japan.
Organizational Affiliation: