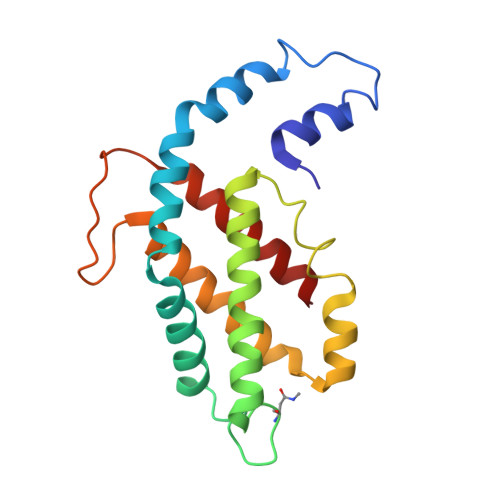
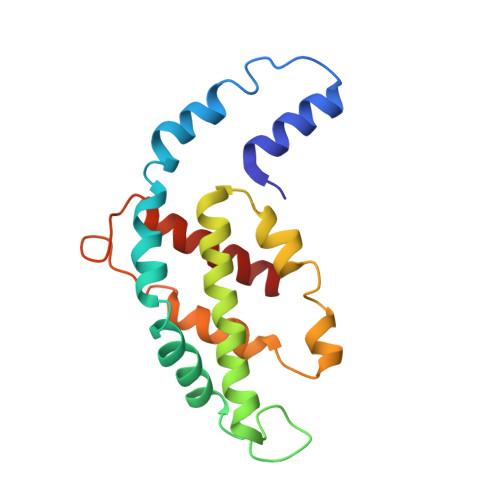
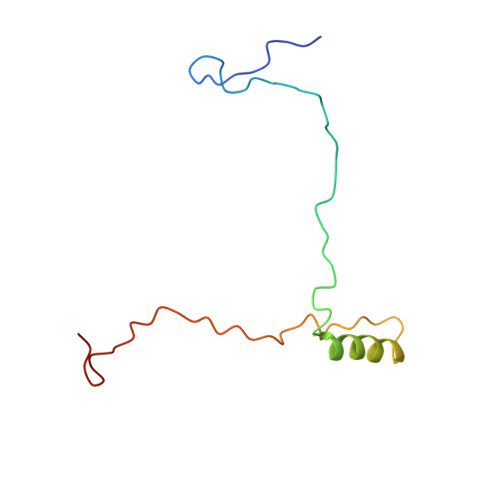
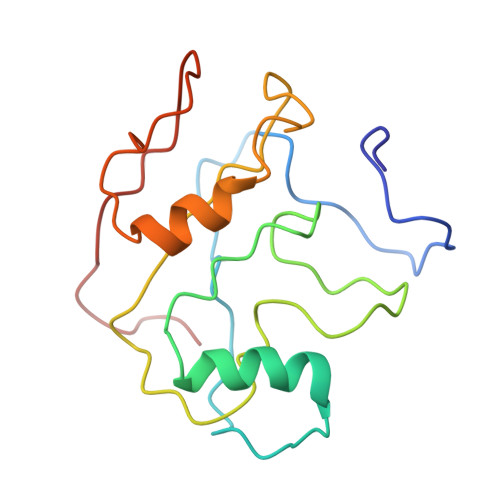
In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
You, X., Zhang, X., Cheng, J., Xiao, Y., Ma, J., Sun, S., Zhang, X., Wang, H.W., Sui, S.F.(2023) Nature 616: 199-206
- PubMed: 36922595
- DOI: https://doi.org/10.1038/s41586-023-05831-0
- Primary Citation of Related Structures:
7Y1A, 7Y4L, 7Y5E, 7Y7A - PubMed Abstract:
In oxygenic photosynthetic organisms, light energy is captured by antenna systems and transferred to photosystem II (PSII) and photosystem I (PSI) to drive photosynthesis 1,2 . The antenna systems of red algae consist of soluble phycobilisomes (PBSs) and transmembrane light-harvesting complexes (LHCs) 3 . Excitation energy transfer pathways from PBS to photosystems remain unclear owing to the lack of structural information. Here we present in situ structures of PBS-PSII-PSI-LHC megacomplexes from the red alga Porphyridium purpureum at near-atomic resolution using cryogenic electron tomography and in situ single-particle analysis 4 , providing interaction details between PBS, PSII and PSI. The structures reveal several unidentified and incomplete proteins and their roles in the assembly of the megacomplex, as well as a huge and sophisticated pigment network. This work provides a solid structural basis for unravelling the mechanisms of PBS-PSII-PSI-LHC megacomplex assembly, efficient energy transfer from PBS to the two photosystems, and regulation of energy distribution between PSII and PSI.
- State Key Laboratory of Membrane Biology, Beijing Frontier Research Center for Biological Structures, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing, China.
Organizational Affiliation: